Abstract
The Mycalesis intermedia belongs to Nymphalidae in Lepidoptera. We described the complete mitogenome of M. intermedia, which is typical circular duplex molecules and 15,386 bp in length, containing the standard metazoan set of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and an A + T-rich region with macro-repeat sequences. All the inferred tRNA secondary structures show the common cloverleaf pattern, with the exception of trnS1(AGN) which lacks the DHU arm. The M. intermedia mitochondrial genome has the same gene order with other lepidopterans.
The Mycalesis intermedia Moore [1892] is a forestry pest, and widely distributed in world (Mayekar and Kodandaramaiah Citation2017). In this study, We sequenced and described the complete mitogenome of M. intermedia to provide useful genetic information for species identification and phylogenetic analysis (Lu et al. Citation2013). The specimens of M. intermedia a were collected by light trapping in Taiyuan, China (37.833393, 112.666114) in July 2018, some of these specimens were immediately frozen in −80 °C on board for mitogenome analysis, and others were preserved by spreading wings in the Herbarium of Institute of Plant Protection, Shanxi Academy of Agricultural Sciences, and their numbers are TYKD20190601-20190604.
The complete mitogenome sequence of M. intermedia is 15386 bp (GenBank accession number: MN610565). As with other insect mitogenomes, the major strand encodes a larger number of genes (9 PCGs and 14 tRNAs) than the minor strand (4 PCGs, 8 tRNAs and 2 rRNA genes). A large non-coding, A + T-rich region in insects is present between rrnS and trnM . The mitochondrial gene order of M. intermedia is identical to those of all other sequenced lepidopterans (Hu et al. Citation2010).
Twelve PCGs have the usual start codon ATN, but the cox1 gene commences with exceptional codon CGA, which was found in another insects as the initial codon (Wu, Zhao, Su, Luo, et al. Citation2016). Twelve PCGs have the common stop codon TAA, except for nad4 have the in-complete stop codon T.
The M. intermedia mitogenome is biased toward A + T (80.95%). The overall base composition is 39.24% A, 41.62% T, 7.32% G and 11.82% C. The 22 tRNA genes ranged from 60 to 70 nucleotides. Fourteen tRNAs are coded on the J-strand and others on the N-strand. Complete cloverleaf secondary structures could be inferred for 21 of the 22 tRNAs. The secondary structure of trnS1(AGN) is incomplete, lacking the DHU arm. The rrnL gene (1375 bp) is located between trnL(CUN) and trnV, and the rrnS (770 bp) between trnV and the A + T-rich region.
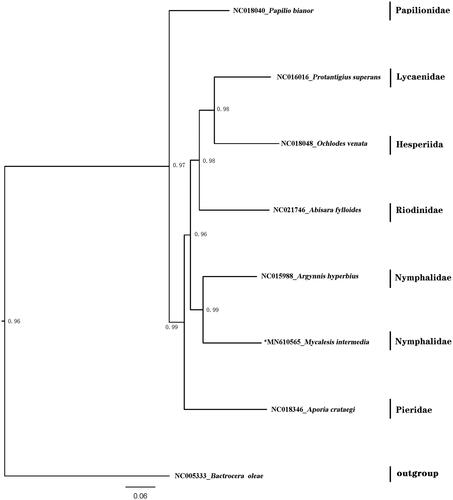
The mitogenome includes 14 overlapping regions ranging from 2 bp to 55 bp and 17 intergenic spacers ranging from 1 bp to 35 bp, with the exception of the A + T-rich region (the largest non-coding region). The A + T-rich region (512 bp) is known for regulating transcription and replication of the mitogenome (Wu, Zhao, Su, He, et al. Citation2016). There is a motif ATAGA in downstream of rrnS followed by an 19 bp, and after 81 bp interval there is another 18 bp Poly-T stretch. The phylogenetic relationships were constructed based on the complete mitochondrial genomes of butterflies represented eight Family of Lepidoptera (). The sequences were aligned with MAFFT v7.2 software (Katoh and Standley Citation2013) and the evolutionary analyses were conducted with RAxML v8.2.10 (Stamatakis Citation2014) on the CIPRES Science Gateway (Miller et al. Citation2010). The GTRGAMMA model with ‘Let RAxML halt bootstrapping automatically’ was used. The phylogenetic tree was visualized using FigTree v1.4.4 (FigTree Citation2018).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- FigTree - molecular evolution, phylogenetics and epidemiology. 2018. [accessed 2018 Nov 26]. http://tree.bio.ed.ac.uk/software/figtree/.
- Hu J, Zhang DX, Hao JS, Huang DY, Cameron S, Zhu CD. 2010. The complete mitochondrial genome of the yellow coaster, Acraea issoria (Lepidoptera: Nymphalidae: Heliconiinae: Acraeini): sequence, gene organization and a unique tRNA translocation event. Mol Biol Rep. 37(7):3431–3438.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lu HF, Su TJ, Luo AR, Zhu CD, Wu CS. 2013. Characterization of the complete mitochondrion genome of diurnal moth Amata emma (Butler) (Lepidoptera: Erebidae) and its phylogenetic implications. PloS One. 8(9):e72410.
- Mayekar HV, Kodandaramaiah U. 2017. Pupal colour plasticity in a tropical butterfly, Mycalesis mineus (Nymphalidae: Satyrinae). PLoS One. 12(2):e0171482.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14; New Orleans, Louisiana: Institute of Electrical and Electronics Engineers (IEEE); p. 1–8.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu YP, Zhao JL, Su TJ, Luo AR, Zhu CD. 2016. The complete mitochondrial genome of Choristoneura longicellana (Lepidoptera: Tortricidae) and phylogenetic analysis of Lepidoptera. Gene. 591(1):161–176.
- Wu YP, Zhao JL, Su TJ, He QS, Xie JL, Zhu CD. 2016. The complete mitochondrial genome of Carposina sasakii (Lepidoptera: Carposinidae). Mitochondrial DNA. 27(2):1432–1434.