Abstract
The complete mitogenome of Thoosa mismalolli Carballo, Cruz-Barraza & Gómez, 2004 (Tetractinellida, Thoosidae) was sequenced. This is the first complete mitogenome of the suborden Thoosina and the third Tetractinellid so far. The mitochondrial genome of T. mismalolli was assembled based on reads obtained with the Illumina HiSeq platform. The length of complete mitogenome is 19,019 bp long and contained 14 protein-coding genes and 23 tRNA, with two tRNA genes. Phylogenetic reconstruction (maximum-likelihood) based on mitogenome of Tetractinellids, supports T. mismalolli as a sister group. This result is congruent with those obtained with molecular markers (CO1, 18S, and 28S), supporting the monophyletic status of Thoosa and providing additional molecular data in favor of the suborder Thoosina.
Introduction
Marine sponges are an abundant and diverse benthic groups that perform relevant functional roles in the marine ecosystems (Bell Citation2008). Boring sponges of genus Thoosa are important because they excavate calcareous substrate, through chemical and physical mechanisms, and therefore contribute with the carbonate budget in these ecosystems (Pomponi Citation1977; Nava and Carballo Citation2008). At the Northeastern Tropical Pacific (NTP), three species of Thoosa have been recorded. Thoosa mismalolli is widespread distributed along the NTP reef communities where is one of the most conspicuous and aggressive bioeroder (Carballo et al. Citation2013). Reproductive biology and ecology of this species is well known (Bautista-Guerrero et al. Citation2016), however, there is no knowledge of its mitogenome which is fundamental to assess the population genetics and its evolutionary biology. This study describes the mitogenome of T. mismalolli, and their phylogenetic relationships with other tetractinellids.
Samples of T. mismalolli invading pocilloporid coral, were collected at Isabel Island, Mexico (21.52°N, 105.54°W). Specimens (voucher: LEB-ICML-UNAM-3179) were deposited in the sponge collection (OAX-MAM-135-10-02) at the Instituto de Ciencias del Mar y Limnología-UNAM. Genomic DNA was extracted using SV Promega kit following the manufacturer’s instruction. Genomic library was prepared using the Nextera® XT DNA library kits, and sequenced with the Illumina HiSeq platform. A total of 15’082,838 paired-reads were obtained, and processed with Trimmomatic (Bolger et al. Citation2014). Only 13,107,511 paired-reads with an average Qscore of 37.9 were retained, and assembled de novo using Megahit v1.2.2-beta (Li et al. Citation2016) and local BLAST. The overlapping ends were trimmed, and the starting position was re-oriented using the mitogenome of Poecillastra laminaris (NCBI: KM362735.1). The annotation was performed in MITOS web server (Bernt et al. Citation2013).
The complete mitogenome of T. mismalolli consist of 19,019 bp (GenBank: MN587873) and included 14 protein-coding genes, 23 tRNA genes and two rRNA subunits. The nucleotide composition is A (29.9%), T (36.8%), C (12.8%) and G (20.4%) with a GC content of 33.2%. The arrangement of protein in the mitogenome of T. mismalolli was similar to the rest of tetractinellid sponges; however, it is important to emphasize that it resulted 4.8% longer than Pocillastra laminaris, ∼3.18% than both Geodia neptuni and Cinachyrella kuekenthali (Lavrov et al. Citation2005, Citation2008; Zeng et al. Citation2016). This change in the mitogenome size is essentially due to insertions in non-coding regions.
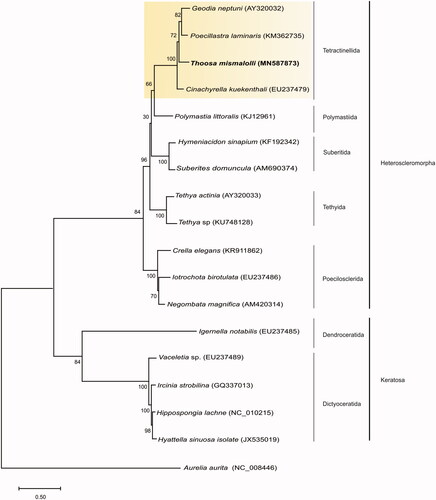
Phylogenetic analysis was performed using MEGA7® software to reconstruct ML tree (Tamura et al. Citation2013). GTR + G resulted as the best-fitting nucleotide substitution model according to the JModelTest 2.0 Software (Posada Citation2008). Aurelia aurita (NC_008446.1) was used as outgroup. Phylogenetic reconstruction () indicated that T. mismalolli is monoplyletic and formed a strongly supported clade with the sister Tetractinellida group. Overall, relationship was congruent with earlier findings on individual mitochondrial (CO1 mtDNA) and ribosomal (28S and 18S rRNA) markers as well as morphological data (Carballo et al. Citation2018), confirming the monophyletic status of Thoosa and supporting the recent creation of new suborder (Thoosina) of Tetractinellida proposed by Carballo et al. (Citation2018).
Figure 1. Consensus tree of Thoosa mismalolli using Maximum-Likelihood (ML) method based on complete mitogenomes of species belonging to subclass Heteroscleromorpha and Verongimorpha. Bootstrap values are indicated above the principal node with 1000 bootstrap replicates. Taxon names are followed by their Genbank accession number. Yellow box indicates the Tetractinellida Order.
Acknowledgements
Special thanks to Bruno Gomez-Gil, for granting access to the BioBacter-CIAD server, and Benjamin Yáñez-Chavéz (ICMyL—Mazatlán) for his assistance in field sampling and data collection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bautista-Guerrero E, Carballo JL, Aguilar-Camacho JM, Sifuentes-Romero I. 2016. Molecular and morphological differentiation of sympatric larvae of coral excavating sponges of genus Thoosa. Zoomorphology. 135(2):159–165.
- Bell JJ. 2008. The functional roles of marine sponges. Estuar Coast Shelf Sci. 79(3):341–353.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Carballo JL, Bautista E, Nava H, Cruz-Barraza JA, Chávez JA. 2013. Boring sponges, an increasing threat for coral reefs affected by bleaching events. Ecol Evol. 3(4):872–886.
- Carballo JL, Bautista-Guerrero E, Cárdenas P, Cruz-Barraza JA, Aguilar-Camacho JM. 2018. Molecular and morphological data from Thoosidae in favour of the creation of a new suborder of Tetractinellida. Syst Biodivers. 16(5):512–521.
- Carballo JL, Cruz-Barraza JA, Gómez P. 2004. Taxonomy and description of clionaid sponges (Hadromerida, Clionaidae) from the Pacific Ocean of Mexico. Zool J Linn Soc-Lond. 141(3):353–397.
- Lavrov DV, Forget L, Kelly MS, Lang BF. 2005. Mitochondrial genomes of two demosponges provide insights into an early stage of animal evolution. Mol Biol Evol. 22(5):1231–1239.
- Lavrov DV, Wang X, Kelly M. 2008. Reconstructing ordinal relationships in the Demospongiae using mitochondrial genomic data. Mol Phylogenet Evol. 49(1):111–124.
- Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, Yamashita H, Lam TW. 2016. MEGAHIT v1. 0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 102:3–11.
- Nava H, Carballo JL. 2008. Chemical and mechanical bioerosion of boring sponges from Mexican Pacific coral reefs. J Exp Biol. 211(17):2827–2831.
- Pomponi S. 1977. Etchings cells of boring sponges: an ultrastructural analysis. Proc. 3rd Intl Coral Reef Symp. 2:486–490.
- Posada D. 2008. ModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Zeng C, Thomas LJ, Kelly M, Gardner J. 2016. The complete mitochondrial genome of the deep-sea sponge Poecillastra laminaris (Astrophorida, Vulcanellidae). Mitochondrial DNA A. 27:1658–1659.
