Abstract
In the present study, the complete sequence of the mitochondrial genome (mitogenome) of Papilio paris (Lepidoptera: Papilionidae) is described. The mitogenome (15,347 bp) of P. paris encodes 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and an adenine (A) + thymine (T)-rich region. Its gene complement and order is similar to that of other sequenced Lepidopterans. Phylogenetic analyses based on 13 PCGs using maximum-likelihood (ML) revealed that P. paris resides in the Papilionoidea family. This study provided the valuable evidence on phylogenetic relationship of the P. paris at the molecular level and essential resource for further research on this species.
Papilio paris (Lepidoptera: Papilionidae) is a butterfly from the Australasia/Indomalaya (Australia) ecozone. This butterfly can be observed from India until China and also from China until Indonesia. Papilio paris is mainly distributed in thickets on slopes and broad-leaved forests, such as rue family plants. The species is one of the important leaf defoliators on Melicope pteleifolia. It has five generations a year in Guangzhou, South China, and the generations overlap. The 1–3 instar larvae mainly feed on the young leaves. After the 3th instar larvae, both young leaves and old leaves are fed. When the density of the insect population is large, the young shoots also are taken. It affects the growth of the plants. Moreover, P. paris is also one of the most ornamental butterflies. To date, there was less information about the complete mitochondrial genomes of P. paris in GenBank. In our study, the complete mitogenome of P. paris was determined, which would be useful for further genetic studies, phylogenetic analysis, and conservation of this species.
The P. paris specimens were collected from Dashushan Forest Park (31°50′53.03″N, 117°11′1.01″E), Anhui Province, China. The collected specimens were identified as P. paris based on the morphological characters by the taxonomist of the Department of Entomology, Anhui Agricultural University, Hefei, China (AHAU). The specimen was stored in 100% ethanol at −80 °C in Bengbu Medical College, Bengbu, China with an accession number: P20160502. The total DNA was extracted using the Genomic DNA Extraction Kit (Aidlab Co., Beijing, China) to the manufacturer instructions. The sequence was amplified using PCR with 12 pairs of primer.
In general, the length of this sequence is 15,347 bp (Genbank No: MN629008), containing two ribosomal RNA genes, 22 putative transfer RNA (tRNA) genes, 13 protein-coding genes, and an adenine (A) + thymine (T)-rich region consisted with previous studies (Sun et al. Citation2016). The 12 PCGs initiated by ATN codons except for cytochrome c oxidase subunit 1 (cox1) gene that is seemingly initiated by the CGA codon as documented in other insect mitogenomes (Dai et al. Citation2015; Sun, Zhang, et al. Citation2017). The mitogenome of P. paris contains 22 tRNAs (ranging from 61 to 71 nucleotides in length) commonly present in most of Lepidopteran mitogenomes. The 16S rRNA (1325 bp) and 12S rRNA (767 bp) were separated by tRNAVal (Wu et al. Citation2013). The A + T-rich region is 533 bp long and contains some conserved regions, including ‘ATAGA’ motif followed by a 11 bp poly-T stretch and a microsatellite-like element (AT)8. The length of poly-T stretch varies from species to species(Dai et al. Citation2015; Sun Citation2019), whereas ATAGA region is conserved in Lepidoptera species (Sun, Chen, et al. Citation2017).
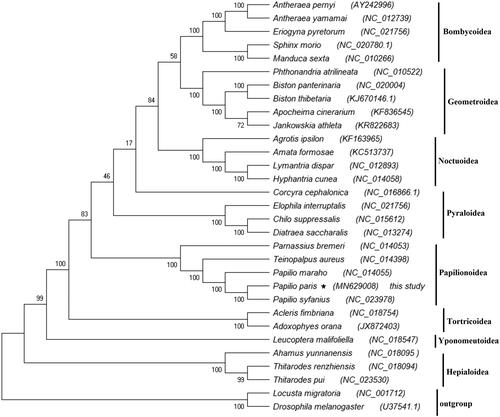
To reconstruct the phylogenetic relationship among Lepidopteran insects, the nucleotide sequences of the 13 PCGs were first aligned and then concatenated; 31 complete mitogenomes were downloaded from the GenBank database. We reconstructed the phylogenetic relationships using Maximum-Likelihood (ML) method based on concatenated nucleotide sequences of the 13 PCGs of the related Lepidopteran superfamilies. The mitogenomes of Drosophila melanogaster (U37541.1) (Lewis et al. Citation1995) and Locusta migratoria (NC_001712) (Flook et al. Citation1995) were used as outgroup. The multiple alignments of the 13 PCGs concatenated nucleotide sequences were conducted using ClustalX version 2.0. (Thompson et al. Citation1997). Then, a concatenated set of nucleotide sequences from the 13 PCGs was used for phylogenetic analyses, which were performed using the ML method with the MEGA X program. The phylogenetic analysis reveals that Bombycoidea, Noctuoidea, Geometroidea, Pyraloidea, Papilionoidea, Tortricoidea, Yponomeutoidea, and Hepialoidea families are monophyletic, and Pyraloidea is more closely related to the Papilionoidea superfamiliy. The different species of same family forms a single cluster; moreover, P. paris was found closely related to P. syfanius of the Papilionoidea (). The results are consistent with previous studies. It is concluded from the results that further studies are required to confirm the phylogentic relationships among these superfamiles.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dai L, Qian C, Zhang C, Wang L, Wei G, Li J, Zhu B, Liu C. 2015. Characterization of the complete mitochondrial genome of Cerura menciana and comparison with other Lepidopteran insects. PLoS One. 10(8):e0132951.
- Flook PK, Rowell CH, Gellissen G. 1995. The sequence, organization, and evolution of the Locusta migratoria mitochondrial genome. J Mol Evol. 41(6):928–941.
- Lewis DL, Farr CL, Kaguni LS. 1995. Drosophila melanogaster mitochondrial DNA: completion of the nucleotide sequence and evolutionary comparisons. Insect Mol Biol. 4(4):263–278.
- Sun Y, Chen C, Gao J, Abbas MN, Kausar S, Qian C, Wang L, Wei G, Zhu BJ, Liu CL. 2017. Comparative mitochondrial genome analysis of Daphnis nerii and other Lepidopteran insects reveals conserved mitochondrial genome organization and phylogenetic relationships. PLoS One. 12(6):e0178773.
- Sun Y, Tian S, Qian C, Sun Y-X, Abbas M N, Kausar S, Wang L, Wei G, Zhu B-J, Liu C-L. 2016. Characterization of the complete mitochondrial genome of Spilarctia robusta (Lepidoptera: Noctuoidea: Erebidae) and its phylogenetic implications. Eur J Entomol. 113:558–570.
- Sun Y, Zhang J, Li Q, Liang D, Abbas MN, Qian C, Wang L, Wei G, Zhu BJ, Liu CL. 2017. Mitochondrial genome of Abraxas suspecta (Lepidoptera: Geometridae) and comparative analysis with other Lepidopterans. Zootaxa. 4254(5):501–519.
- Sun Y, Zhang X, Liu S, Xia J, Geng J, Zhang K. 2019. The complete mitochondrial genome of the Parasa tessellata (Lepidoptera: Limacodidae). Mitochondrial DNA B. 4(1):1690–1691.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882.
- Wu QL, Liu W, Shi BC, Gu Y, Wei SJ. 2013. The complete mitochondrial genome of the summer fruit tortrix moth Adoxophyes orana (Lepidoptera: Tortricidae). Mitochondrial DNA. 24(3):214–216.