Abstract
In the present study, the complete mitochondrial genome of T. szidati was assembled by next generation sequencing (NGS). We found that the complete mitochondrial genome of T.szidati is 14, 303 bp in length and consists of 3023 (21.1%) adenine, 1153 (8.1%) cytosine, 3432 (24.0%) guanosine and 6695 (46.8%) thymine. The genome contains 12 conserved core protein-coding genes (atp6, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, CYTB), 21 tRNA genes, 2 rRNA genes and 1 D-loop region. Phylogenetic analysis showed that T. szidati has a close relationship with T. regent. Knowledge of mitochondrial genome of T. szidati could provide useful information for the further studies of evolutionary biology, epidemiology and species identification.
Trichobilharzia is a kind of thread-like schistosome and it is distributed in birds worldwide (Yakhchali et al. Citation2016). It was reported that there were over 30 species in this kind of schistosomes (Loker Citation2009). T. szidati is one of the species that belongs to Schistosomatidae, Trematoda (Brant and Loker Citation2009). T. szidati can only parasitize in specific birds hosts whereas its larvae could penetrate, transform and migrate in all kinds of hosts (Le et al. Citation2002). In freshwater, the larva of T. szidati has the ability to penetrate human skin. Besides, it may cause human cercarial dermatitis (Martin Kašný Citation2011). Adult worms of Trichobilharzia cause extensive damage as they migrate and lay larvae in their preferred site (either the nasal mucosa or visceral capillaries) of infection (Zbikowska Citation2005). In the present study, the mitochondrial genome of T. szidati from China was sequenced. Its genetic characteristics and phylogenetic status was assessed in order to provide useful information for further study on trematode evolution.
Trichobilharzia szidati cercariae were collected from host snail Lymnaea stagnalis in Henan, China. (34°40′E; 112°21′N) and was stored in Henan Agricultural University (No. Tsz002). Total genomic DNA was isolated from this specimen according to Webster et al (Webster et al. Citation2007), and was stored in the sequencing company (BGI Tech, Shenzhen, China). We constructed sequencing libraries using a NEB Next Ultra II DNA Library Prep Kit (NEB, Beijing, China) following the manufacturer’s instructions. Whole genomic sequencing was performed using an Illumina HiSeq 2500 Platform (Illumina, San Diego, CA, USA). The mitogenome was assembled and annotated as reported (Li, Liao, et al. Citation2018; Li, Wang, et al. Citation2018; Li et al. Citation2019; Li et al. Citation2019a).
The complete mitochondrial genome of T. szidati is 14, 303 bp in length and consists of 3023 (21.1%) adenine, 1153 (8.1%) cytosine, 3432 (24.0%) guanosine and 6695 (46.8%) thymine. The genome contains 12 conserved core protein-coding genes (atp6, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, CYTB), 21 tRNA genes, 2 rRNA genes and 1 D-loop region. The T. szidati mitochondrial genome sequence was submitted to GenBank under the accession number of MG570047.
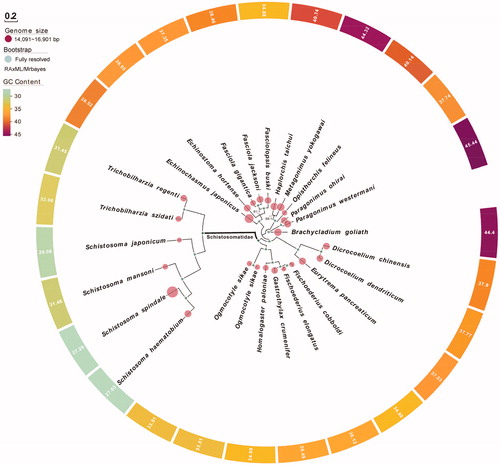
We used maximum likelihood (ML) and Bayesian inference (BI) to create phylogenies based on the combined gene alignment (Li, Yang, et al. Citation2018; Li et al. Citation2019b; Li, Ren, et al. Citation2019). The ML analysis was performed with RAxML (Stamatakis Citation2014), while bootstrap values were calculated using 1,000 replicates to assess node support (Li, He, et al. Citation2020). Bayesian analyses were performed with MrBayes v3.2.6 (Ronquist et al. Citation2012). Phylogenetic analysis showed that T. szidati has a close relationship with T. regent (Semyenova et al. Citation2017) ().
Figure 1. Phylogenetic relationships of 26 digeneans based on Bayesian inference and maximum likelihood analysis of 12 protein-coding genes. Support values are bootstrap values (before slash) and bayesian posterior probabilities (after slash). Brachycladium goliath (KR703278); Dicrocoelium chinensis (KF318786); Dicrocoelium dendriticum (KF318787); Echinochasmus japonicas (KP844722); Echinostoma hortense (KR062182); Eurytrema pancreaticum (KP241855); Fasciola gigantica (KF543342); Fasciola jacksoni (KX787886); Fasciolopsis buski (KX169163); Fischoederius cobboldi (KX169164); Fischoederius elongates (KM397348); Gastrothylax crumenifer (KM400624); Haplorchis taichui (KF214770); Homalogaster paloniae (KT266674); Metagonimus yokogawai (KC330755); Ogmocotyle sikae 1 (KR006934); Ogmocotyle sikae 2 (NC_027112); Opisthorchis felineus (EU921260.2); Paragonimus ohirai (KX765277); Paragonimus westermani (KX943544); Schistosoma haematobium (DQ157222.2); Schistosoma japonicum (JQ781206); Schistosoma mansoni (NC_002545); Schistosoma spindale (DQ157223); Trichobilharzia regent (DQ859919).
Disclosure statement
The authors have declared that no competing interests exist.
Additional information
Funding
References
- Brant SV, Loker ES. 2009. Molecular systematics of the avian schistosome genus Trichobilharzia (Trematoda: Schistosomatidae) in North America. J Parasitol. 95(4):941–963.
- Kašný M, Mikeš L, Doleckova K, Hampl V, Dvorak J, Novotný M, Horak P. 2011. Cathepsins B1 and B2 OF Trichobilharzia SPP., bird schistosomes causing cercarial dermatitis. Adv Exp Med Biol. 712:136–154.
- Le TH, Blair D, McManus DP. 2002. Mitochondrial genomes of parasitic flatworms. Trends Parasitol. 18(5):7.
- Li Q, He X, Ren Y, Xiong C, Jin X, Peng L, Huang W. 2020. Comparative mitogenome analysis reveals mitochondrial genome differentiation in ectomycorrhizal and asymbiotic Amanita species. Front Microbiol. doi: 10.3389/fmicb.2020.01382.
- Li Q, Liao M, Yang M, Xiong C, Jin X, Chen Z, Huang W. 2018. Characterization of the mitochondrial genomes of three species in the ectomycorrhizal genus Cantharellus and phylogeny of Agaricomycetes. Int J Biol Macromol. 118(Pt A):756–769.
- Li Q, Ren Y, Shi X, Peng L, Zhao J, Song Y, Zhao G. 2019. Comparative Mitochondrial Genome Analysis of Two Ectomycorrhizal Fungi (Rhizopogon) Reveals Dynamic Changes of Intron and Phylogenetic Relationships of the Subphylum Agaricomycotina. Int J Mol Sci. 20(20):5167.
- Li Q, Wang Q, Chen C, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2018. Characterization and comparative mitogenomic analysis of six newly sequenced mitochondrial genomes from ectomycorrhizal fungi (Russula) and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 119:792–802.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Liu Q, Huang W. 2019. Characterization and comparative analysis of six complete mitochondrial genomes from ectomycorrhizal fungi of the Lactarius genus and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 121:249–260.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2019a. Characterization and comparison of the mitochondrial genomes from two Lyophyllum fungal species and insights into phylogeny of Agaricomycetes. Int J Biol Macromol. 121:364–372.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2019b. The first complete mitochondrial genome from the family Hygrophoraceae (Hygrophorus russula) by next-generation sequencing and phylogenetic implications. International Journal of Biological Macromolecules. 122:1313–1320.
- Li Q, Yang M, Chen C, Xiong C, Jin X, Pu Z, Huang W. 2018. Characterization and phylogenetic analysis of the complete mitochondrial genome of the medicinal fungus Laetiporus sulphureus. Sci Rep. 8(1):9104.
- Loker S. 2009. Molecular systematics of the avian schistosome genus Trichobilharzia (Trematoda: Schistosomatidae) in North America. J Parasitol. 95(4):941–963.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 61(3):539–542.
- Semyenova S, Chrisanfova G, Mozharovskaya L, Guliaev A, Ryskov A. 2017. The complete mitochondrial genome of the causative agent of the human cercarial dermatitis, the visceral bird schistosome species Trichobilharzia szidati (platyhelminthes: Trematoda: Schistosomatidae). Mitochondrial DNA Part B. 2(2):469–470.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313.
- Webster BL, Rudolfova J, Horak P, Littlewood DT. 2007. The complete mitochondrial genome of the bird schistosome Trichobilharzia regenti (Platyhelminthes: Digenea), causative agent of cercarial dermatitis. J Parasitol. 93(3):553–561.
- Yakhchali M, Hosseinpanahi A, Malekzadeh-Viayeh R. 2016. Molecular evidence of trichobilharzia species (Digenea: Schistosomatidae) in the snails of Lymnaea auricularia from Urmia Suburb, North West Iran. Iran J Parasitol. 11(3):296–302.
- Zbikowska E. 2005. Do larvae of Trichobilharzia szidati and echinostoma revolutum generate behavioral fever in Lymnaea stagnalis individuals? Parasitol Res. 97(1):68–72.
