Abstract
Trophis caucana, which belongs to Moraceae, is a tree species lived in a humid climate at low and middle altitudes. The complete chloroplast (cp) genome of T. caucana was sequenced and assembled in this study. The cp genome is 161,445 bp in length with comprising two copies of inverted region (IR, 25,894 bp) separated by the large single copy (LSC, 89,633 bp) and small single copy (SSC, 20,024 bp) regions. It encodes 111 unique genes, consisting of 77 protein-coding genes, 30 tRNA genes, and four rRNA genes, with 19 duplicated genes in the IR regions. Phylogenetic analysis indicates that T. caucana is sister to Antiaris toxicaria in Moraceae family.
Trophis P. Browne is a small genus in the plant family of Moraceae which includes about nine species with six distributed in Neotropical and three are Palaeotropical. The trees of this genus have alternate leaves, small dioecious green flowers in usually spicate or racemose clusters, and a nearly round thin-fleshed fruit with a single rather large seed (Berg Citation1988). Previous studies have proved that the phylogeny of this genus was especially problematic, and the species within the genus was polyphyletic in phylogenetic tree based on ndhF gene (Datwyler and Weiblen Citation2004). In the present study, the complete chloroplast genome of T. caucana was first sequenced using genome skimming data. The genome sequence was registered into GenBank with the accession number MN868948.
Total genomic DNA was extracted from silica-dried leaves of one T. caucana plant collected in Abel Iturralde (Bolivia; 67°58′15.03″W, 14°20′57.10″S) using Plant DNAzol Reagent (LifeFeng, Shanghai) according to the manufacturer’s protocol. A voucher specimen (Fuentes Claros and Alfredo Fernando 5323) was deposited at Missouri Botanical Garden Herbarium. High quality DNA was sheared and the paired-end library (≤800 bp) was sequenced on an Illumina HiSeq X10 at Beijing Genomics Institute (BGI, Wuhan, China). The raw data were screened by quality with Phred score <30 and assembled into contigs using the CLC Genomic Workbench (CLC Inc. Aarhus, Denmark). The complete cp genome of T. caucana was reconstructed with Cannabis sativa (GenBank accession number: KP274871) as a reference and annotated using the software Geneious R11 (Biomatters, Auckland, New Zealand) following description in Liu et al. (Citation2017, Citation2018). We drew the circular cp genome map of T. caucana using the OGDRWA program (Lohse et al. Citation2013). Phylogenetic tree for 24 whole cp genome sequences of Rosales was constructed using maximum-likelihood (ML) method implemented in RAxML-HPC v8.1.11 on the CIPRES cluster (Miller et al. Citation2010) with Lagenaria sceraria and Morella rubra as outgroups.
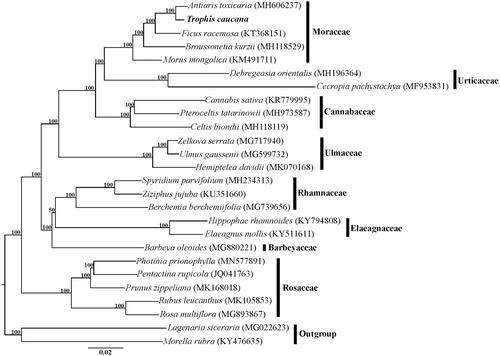
The cp genome of T. caucana was 161,445 bp in length and had a typical quadripartite structure consisting of an 89,633 bp large single copy region (LSC), a 20,024 bp small copy region (SSC) and two 25,894 bp inverted repeats (IRs). Within the genome of T. caucana, there are 111 unique genes, including 77 protein-coding genes, 30 tRNA genes, and four rRNA genes, additionally with 19 duplicated genes in the IR regions. Among the 111 genes, six tRNA genes and eight protein-coding genes contain a single intron, and three genes (rps12, clpP, and ycf3) contain two introns. The overall GC content of the total length, LSC, SSC, and IR regions is 35.7%, 33.3%, 28.8%, and 42.6%, respectively. The phylogeny revealed that the five representative genera of Moraceae, including Antiaris, Trophis, Ficus, Broussonetia, and Morus, formed a highly supported clade, and T. caucana is sister to A. toxicaria within this family ().
Figure 1. Phylogenetic relationships of Rosales inferred based on whole chloroplast genome sequences. Numbers above the branches represent bootstrap values from maximum-likelihood analyses.
In conclusion, the complete cp genome of T. caucana is reported for the first time in this study. It will provide essential and important genetic resources for future better investigation of phylogeny within the genus Trophis.
Acknowledgments
We sincerely thank the DNA bank of Missouri Botanical Garden for providing the DNA sample.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Berg CC. 1988. The genera Trophis and Streblus (Moraceae) remodeled. Proceedings of the Koninklijke Nederlandse Akademie Van Wetenschappen, Amsterdam, The Netherlands, Series C 91; p. 345–362.
- Datwyler SL, Weiblen GD. 2004. On the origin of the fig: phylogenetic relationships of Moraceae from ndhF sequences. Am J Bot. 91(5):767–777.
- Liu LX, Li R, Worth JR, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968
- Liu LX, Li P, Zhang HW, Worth J. 2018. Whole chloroplast genome sequences of the Japanese hemlocks, Tsuga diversifolia and T. sieboldii, and development of chloroplast microsatellite markers applicable to East Asian Tsuga. J Forest Res. 23(5):318–323.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE). 2010 IEEE; p. 1–8.
