Abstract
Sparassis crispa, also known as cauliflower mushroom, is a widely used medicinal mushroom in traditional Chinese medicine due to the presence of bioactive substances with pharmacological activity. Here, we report a complete mitochondrial genome sequence of S. crispa consisting of 139,253 bp containing 47 genes including 15 protein-coding genes, 27 transfer RNA, and 5 ribosomal RNA genes obtained from 40.406 Mb genome containing 18,917 predicted contigs using raw data of next-generation sequencing having 85.4% Q30. The overall base composition of S. crispa was 26.47% G-C and 73.53% A-T. The phylogenetic tree based on atp6 sequence data showed its close relationship with Sparassis radicata. The complete mitochondrial genome sequence of S. crispa provides an essential and important DNA molecular data for further phylogenetic and evolutionary analysis of S. crispa.
The brown-rot producing cauliflower mushroom, Sparassis crispa, is widely distributed in Australia, North America, Europe, and East Asia (Martin and Gillbertson Citation1976). Sparassis crispa mainly grows on coniferous trees such as Larix kaempferi, Pinus densiflora, Pinus koraiensis (Lee et al. Citation2015). It is known for its medicinal significance due to the availability of various pharmacological substances and their use in health supplements (Kimura Citation2013; Bashir and Choi Citation2017). Previous studies have reported the phylogenetic analyses of mushrooms using nucleotide sequence data from ribosomal DNAs, partial RNA polymerase subunit II gene, and mitochondrial ribosomal DNA (Dai et al. Citation2006; Ryoo et al. Citation2013; Kiyama et al. Citation2018; Xiao et al. Citation2018). Among the eight reported clades in Sparassis spp. [Sparassis brevipes, Sparassis crispa, Sparassis cystidiosa, Sparassis latifolia, Sparassis miniensis, Sparassis radicata, Sparassis spathulate, Sparassis subalpina], the Asian collections of S. crispa are morphologically different from the European collections; thus it is important to identify this mushroom at the level of mitochondrial DNA.
Here, we report the complete mitochondrial genome of S. crispa using next-generation sequencing, which will help to better understand the phylogenetic status of S. crispa. Sparassis crispa was obtained from the Culture Collection of Wild Mushrooms, Incheon, Republic of Korea (37.46 latitude, 126.67 longitude). This specimen is stored at the Culture Collection of Wild Mushrooms, Incheon, Republic of Korea [Reference No. IUM04050]. The mycelium was grown in the laboratory as described previously by Lee et al. (Citation2015) with slight modifications. Briefly, the fruiting bodies of S. crispa were sterilized, isolated, and cultured at 23 ± 2 °C on potato dextrose agar medium. The mitochondrial DNA was sequenced using Illumina Hiseq X sequencing (Illumina Citation2014). The genome assembly was done using CLC assembly 5.1.1 (Qiagen, Aarthus C, Denmark) and the annotation of full sequence was performed using BLAST, tRNAscan-SE2.0 (Lowe and Chan Citation2016), and MITOS webserver (Bernt et al. Citation2013; Han et al. Citation2018).
Length of the complete mitochondrial genome of S. crispa was 139,253 bp containing 47 genes including 27 transfer RNA genes, 15 protein-coding genes of seven NADH dehydrogenases (nad1, and2, nad3, nad4, nad4L, nad5, and nad6), three ATPases (atp6, atp8, and atp9), three cytochrome c oxidases (cox1, cox2, and cox3), an apocytochrome b (cob), and a ribosomal protein S3 (rps3), and 5 ribosomal RNA genes (three rns and two rnl), obtained from 40.406 Mb genome containing 18,917 predicted contigs using raw data of next-generation sequencing having 85.4% Q30 (ratio of bases having Phred quality score of >30). The complete and annotated mitochondrial genome of S. crispa has been deposited to GenBank [accession number: MN722635].
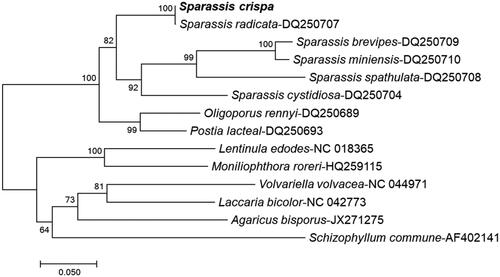
To predict the phylogenetic evaluation of S. crispa, a neighbor-joining phylogenetic tree was constructed with MEGA7 (Tamura et al. Citation2013), using atp6 of representative species belonging to Ascomycota or Basidiomycota, especially, S. radicata (). The phylogenetic analysis with other fungal genomes did not show much difference, suggesting that the previously reported S. radicata (Dai et al. Citation2006) and currently reported S. crispa are very close to each other.
Acknowledgments
This work was supported by the BB21+ Project in 2019.
Disclosure statement
The authors declare that there is no potential conflict of interest regarding the publication of this article.
References
- Bashir KMI, Choi J-S. 2017. Clinical and physiological perspectives of β-glucans: the past, present, and future. IJMS. 18(9):1906.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dai YC, Wang Z, Binder M, Hibbett DS. 2006. Phylogeny and a new species of Sparassis (Polyporales, Basidiomycota): evidence from mitochondrial atp6, nuclear rDNA and rpb2 genes. Mycologia. 98(4):584–592.
- Han J-G, Oh J, Jo JW, Kim CS, Kwag Y-N, Han S-K, Sung G-H. 2018. The complete mitochondrial genome of Sanghuangporus sanghuang (Hymenochaetaceae, Basidiomycota). Mitochondr DNA B. 3(1):456–457.
- Illumina. 2014. Preliminary system specification sheet for human whole-genome sequencing with the HiseqXTM sequencing system; [accessed 26 November 2019]. https://www.illumina.com/content/dam/illumina-marketing/documents/products/appnotes/appnote-hiseq-x.pdf.
- Kimura T. 2013. Natural products and biological activity of the pharmacologically active cauliflower mushroom Sparassis crispa. BioMed Res Int. 2013:1–9.
- Kiyama R, Furutani Y, Kawaguchi K, Nakanishi T. 2018. Genome sequence of the cauliflower mushroom Sparassis crispa (Hanabiratake) and its association with beneficial usage. Sci Rep. 8(1):16053.
- Lee DJ, Jang MC, Jo AR, Choi HJ, Kim K-S, Chi Y-T. 2015. Noble strain of Sparassis latifolia produces high content of β-glucan. Asia Pac J Trop Biomed. 5(8):629–635.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Martin K. J, Gilbertson R. L. 1976. Cultural and other morphological studies of Sparassis radicata and related species. Mycologia. 68(3):622–639.
- Ryoo R, Sou HD, Ka KH, Park H. 2013. Phylogenetic relationships of Korean Sparassis latifolia based on morphological and ITS rDNA characteristics. J Microbiol. 51(1):43–48.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Xiao D, Ma L, Yang C, Ying Z, Jiang X, Lin Y-Q. 2018. De Novo sequencing of a Sparassis latifolia genome and its associated comparative analyses. Can J Infect Dis Med Microbiol. 2018:1–12.