Abstract
Ulva meridionalis, a green macroalgae, is one of the causal species for green tides in Japan and spread into the coast of China. During this research, we sequenced the complete mitochondrial genome of U. meridionalis. The mitogenome is 62,887 bp in length, including 28 encoding genes and 29 tRNA genes. Compared with the Ulva species from mitogenome, the gene order and organization of this mitogenome are similar to most of other determined Ulva mitogenomes, with the nucleotide base composition of A 33.6%, T 32.2%, C 16.2%, and G 18.0%. Phylogenetic analysis shows U. meridionalis is closely related to Ulva flexuosa.
Species causing large-scale green tide disasters worldwide are mainly belong to the genus of Ulva (Blomster et al. Citation2002; Smetacek and Zingone Citation2013; Zhang et al. Citation2019). The classification of Ulva is complex and has been a research hotspot (Ayden et al. Citation2003). Ulva meridionalis was a large marine tropical macroalgae that mainly distributed in southern Japan (Shimada et al. Citation2008; Horimoto et al. Citation2011). This species was never observed in the Yellow Sea of China before. However, in 2019, we discovered that this species widely distributed in the crab aquaculture ponds near the estuary of Dagu river, Jiaozhou-bay area of Qingdao, China (36°12′26.316″N, 120°06′37.756″E), and the specimen was stored in the herbarium of Shanghai Ocean University Museum (SHOU2019QD082001). This species had a strong ecological adaptability, the maximum growth rate can reach up to 0.796 d−1 under the conditions that nitrogen and organic phosphorus concentrations are 4.02 mg/L and 0.125 mg/L (Yang Citation2018), which have a high risk to become a new green tide in China, so it is of great significance to sequence the mitochondrial genome to understand its evolutionary relationship.
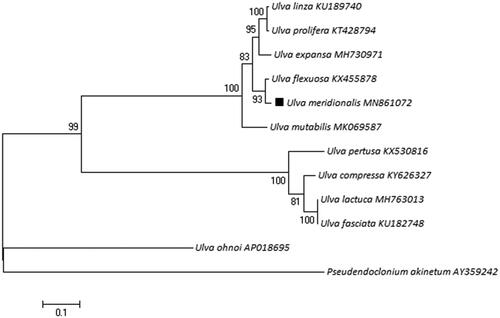
After sample collection, the sample was cultured in the laboratory with VSE medium at 20 degrees Celsius under a light intensity of 100 μmolm−2s−1 (Jiang et al. Citation2019). The complete mitochondrial genome of U. meridionalis is 62,887 bp in length (Genbank accession number: MN861072). The overall base composition of mitochondrial genome is A (33.6%), T (32.2%), C (16.2%), G (18.0%), similar to most of the Ulva macroalgae in mitogenome, and the percentage of A + T (65.8%) is higher than C + G (34.2%). It contains 28 encoding genes and 29 tRNA genes. A Maximum-likelihood (ML) phylogenetic tree with 11 species complete mitochondrial genome of Ulva species and one outgroup (Pseudendoclonium akinetum) was constructed in MEGA 7 software () (Kumar et al. Citation2016), which shows U. meridionalis is closely related to Ulva flexuosa.
Figure 1. The phylogenetic tree relationship of 12 species in Phylum Chlorophyta based on the data of encoding genes. Genbank accession numbers: Ulva meridionalis: MN861072; Ulva compressa: KY626327; Ulva expansa: MH730971; Ulva fasciata: KU182748; Ulva flexuosa: KX455878; Ulva lactuca: MH763013; Ulva linza: KU189740; Ulva mutabilis: MK069587; Ulva ohnoi: AP018695; Ulva pertusa: KX530816; Ulva prolifera: KT428794; Pseudendoclonium akinetum: AY359242.
Acknowledgments
We thank North China Sea Environmental Monitoring Center for providing personnel assistance and vehicle transportation service.
Disclosure statement
We declare there is no apparent or potential conflict of interest in the authors’ report.
Additional information
Funding
References
- Ayden HSH, Aanika J, Aggs AM, Ilva PCS, Aaland J. 2003. Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. Br Phycol Bulletin. 38:277–294.
- Blomster J, Back S, Fewer DP, Kiirikki M, Lehvo A, Maggs CA, Stanhope MJ. 2002. Novel morphology in Enteromorpha (Ulvophyceae) forming green tides. Am J Botany. 89(11):1756–1763.
- Horimoto R, Masakiyo Y, Ichihara K, Shimada S. 2011. Enteromorpha-like Ulva (Ulvophyceae, Chlorophyta) growing in the todoroki river, Ishigaki Island, Japan, with special reference to Ulva meridionalis Horimoto et Shimada, sp. nov. Bull Natl Mus Nat Sci Ser B. 37:155–167.
- Jiang T, Gu K, Wang LK, Liu Q, Shi JT, Liu MM, Tang CY, Su YZ, Zhong SC, Cai CE, et al. 2019. Complete chloroplast genome of Ulva prolifera, the dominant species of green macroalgal blooms in Yellow Sea, China. Mitochondrial DNA Part B. 4(1):1930–1931.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Shimada S, Yokoyama N, Arai S, Hiraoka M. 2008. Phylogeography of the genus Ulva (Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. J Appl Phycol. 20(5):979–989.
- Smetacek V, Zingone A. 2013. Green and golden seaweed tides on the rise. Nature. 504(7478):84–88.
- Yang Y. 2018. Study on nutrient removal from secondary effluent and water purification using Ulva Meridionalis. Xi’an: Shaanxi University of Science and Technology. p. 1–60.
- Zhang YY, He PM, Li HM, Li G, Liu JH, Jiao FL, Zhang JH, Huo YZ, Shi XY, Su RG, et al. 2019. Ulva prolifera green-tide outbreaks and their environmental impact in the Yellow Sea, China. Natl Sci Rev. 6(4):825–838.
