Abstract
The genus Pyrus, comprising several popular fruit crops worldwide, includes over 30 tree species. Here we determined the complete plastid genome sequence of Pyrus betulaefolia. The plastome consists of 160,184 bp, including a pair of inverted repeats (IRs) with a length of 26,384 bp separated by a large single-copy region (LSC) and a small single-copy region (SSC) of 88,121 bp and 19,295 bp, respectively. Further phylogenetic analyze was conducted using 11 complete plastid genomes of Rosaceae with KVM + F + I model, which supports Pyrus betulaefolia as a sister to all other eight Pyrus taxa with published plastomes.
Keywords:
Pyrus betulaefolia Bunge is a subtropical tree that mainly distributed in northern China (http://foc.iplant.cn/). Zong et al. (Citation2013) suggest that its developed root system has a strong ability for P. betulaefolia in cold resistance, drought resistance, and saline-alkali tolerance. It was also presumably considered as one of the ancient species of the genus Pyrus (Rubtsov Citation1944; Zheng et al. Citation2011). P. betulaefolia has good grafting compatibility with other species of Pyrus genus, which is mainly used as the rootstock of all kinds of cultivated pear and is also an important parent in pear dwarfing rootstock and resistance breeding (Okubo and Sakuratani Citation2000; Robbani et al. Citation2006). For a better understanding of the relationships of P. betulaefolia and other Pyrus species, it is necessary to reconstruct a phylogenetic tree based on high-throughput sequencing approaches.
Young leaves of P. betulaefolia were collected from the Ruili Botanical Garden (Long. 97.8185 E, Lat. 24.0714N, 1165m) for genomic DNA extraction using the N-Lauroylsarcosine sodium salt method (Wu et al. Citation2017). The voucher was deposited at the Key Laboratory of Forest Resources Conservation and Utilization in the Southwest Mountains of China Ministry of Education, Southwest Forestry University (Accession Number: SWFU–SY36748). The whole plastome was sequenced following Zhang et al. (Citation2016), and the long-range PCR was used for next-generation sequencing with 15 pairs of universal primers. The contigs were aligned using the publicly available plastid genome of P. ussuriensis (Accession Number: MK172841) (Gil et al. Citation2019) and annotated in Geneious 8.1.9.
The plastome of P. betulaefolia (LAU10003) with a length of 160,184 bp, was the largest of the 25 reported plastome of Pyrus, was 27 bp and 1023 bp larger than that of P. ussuriensis (160,157 bp, MK172841) and P. spinosa (159,161 bp, NC023130). The length of the large single-copy (LSC), inverted repeats (IRs), and small single-copy (SSC) regions of P. betulaefolia was 88,121 bp, 26,384 bp, and 19,295 bp, respectively. The overall GC content is 36.5% (LSC, 34.1%; IR, 42.7%; SSC, 30.3%). The P. betulaefolia plastid genome encoded a set of 133 genes, of which 88 are protein-coding genes, 8 are rRNA genes, and 37 are tRNA genes.
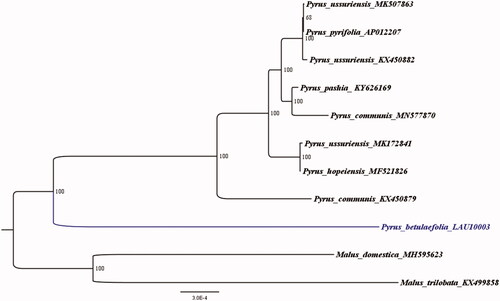
Furthermore, in order to confirm the evolutionary relationship between P. betulaefolia and other species with published plastomes in Pyrus, we reconstructed a phylogenetic tree () based on 10 published plastid genome sequences of the Rosaceae. Malus domestica (Accession Number: MH595623) was treated as an out-group, aligned by the MAFFT version 11 program (Katoh and Standley Citation2013). A maximum-likelihood (ML) analysis based on the KVM + F+I model was performed with iqtree version 1.6.7 program using 1000 bootstrap replicates (Nguyen et al. Citation2015). The ML phylogenetic tree with 68–100% bootstrap values at each node supported that P. betulaefolia as a sister to all other eight Pyrus taxa with published plastomes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability
The chloroplast data of the P. betulaefolia will be submitted to Rosaceae Chloroplast Genome Database (https://lcgdb.wordpress.com). Accession numbers are LAU10003.
Additional information
Funding
References
- Gil HY, Kim YS, Kim SH, Jeon JH, Kwon Y, Kim SC, Park JS. 2019. The complete chloroplast genome of Pyrus ussuriensis Maxim. (Rosaceae). Mitochondr DNA B. 4(1):1000–1001.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Okubo M, Sakuratani T. 2000. Effects of sodium chloride on survival and stem elongation of two Asian pear rootstock seedlings. Sci Hortic. 85(1–2):85–90.
- Robbani M, Banno K, Yamaguchi K, Fujisawa N, Liu JY, Kakegawa M. 2006. Selection of dwarfing pear rootstock clones from Pyrus betulaefolia and P. calleryana seedlings. J Japan Soc Hort Sci. 75(1):1–10.
- Rubtsov GA. 1944. Geographical distribution of the genus Pyrus and trends and factors in its evolution. Am Nat. 78(777):358–366.
- Wu X, Yin H, Chen YY, Qi KJ, Shi DQ, Cao P, Tian YN, Wu J, Zhang SL. 2017. New method for extraction of high quality genomic DNA from pear. South China Fruits. 46(2):31–36.
- Zhang T, Zeng C-X, Yang J-B, Li H-T, Li D-Z. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Syst Evol. 54(3):219–227.
- Zheng XY, Hu CY, Spooner D, Liu J, Cao JS, Teng YW. 2011. Molecular evolution of Adh and LEAFY and the phylogenetic utility of their introns in Pyrus (Rosaceae). BMC Evol Biol. 11(1):255.
- Zong Y, Sun P, Niu QF, Teng YW. 2013. Distribution situation and assessment of morphological diversity of wild Pyrus betulaefolia in Northern China. J Fruit Sci. 30(6):918–923.