Abstract
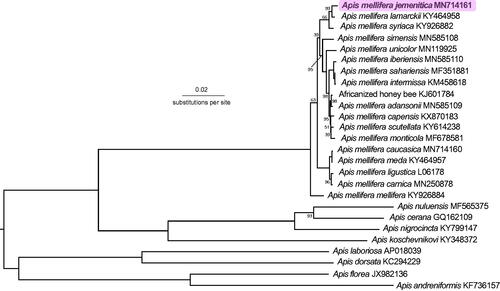
The mitochondrial genome of a worker Apis mellifera jemenitica was 16,623 bp. It consisted of 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs and a control region. Phylogenetic analyses suggest a close relationship between A. m. jemenitica, A. m. lamarckii and A. m. syriaca.
The Arabian honey bee, Apis mellifera jemenitica (Ruttner, 1976, junior synonym: nubica; often written yemenitica), is the only Apis mellifera subspecies to occur naturally in both Africa and Asia with a distribution from the Arabian peninsula in the East, to south of the Sahara in the West (Al-Ghamdi et al. Citation2013). Previous studies on the mitochondrial DNA of A. m. jemenitica are not in agreement as to which A. mellifera lineage this subspecies belongs (e.g. Franck et al. Citation2001; El-Niweiri and Moritz Citation2008; reviewed in Al-Ghamdi et al. Citation2013). Here, we present the mitochondrial genome (GenBank: MN714161) of a worker A. m. jemenitica honey bee (Voucher No. 1544, Ruttner Bee Collection at the Bee Research Institute at Oberursel, Germany). This sample was collected in 1988 by Prof. H. Hoppe in Bani Shiab, Yemen (15°29 N, 43°44 E), and subspecies identity was confirmed morphometrically.
Genomic DNA was extracted and prepared for PE-150 bp sequencing on Illumina Hi-Seq 3000/4000 (San Diego, CA) following the method described by Eimanifar et al. (Citation2017). The quality of the resulting data was assessed using FastQC (Andrews Citation2010), and trimmed with Trimmomatic (Bolger et al. Citation2014). Trimmed data were mapped in Geneious Prime 2019.0.4 (Kearse et al. Citation2012) using the method from Boardman et al. (Citation2019) and the mitogenome of A. m. lamarckii (KY464958) as a reference. Mitos2 was used to annotate the assembled mitogenome (Bernt et al. Citation2013) before it was manually adjusted to match the A. m. capensis annotation (KX870183) in Geneious Prime. The 13 protein-coding genes (PCGs) and two ribosomal RNA (rRNA) sequences were manually aligned to sequences from other Apis mitogenomes in Mesquite v3.5 (Maddison and Maddison Citation2018). For phylogenetic estimation, we used RAxML 8.2.10 (Stamatakis Citation2014) with the GTRGAMMA model and 1000 bootstrap replicates (-f a option) on the CIPRES Science Gateway V.3.3 (Miller et al. Citation2010), and p-distances were obtained using PAUP 4.0a (Swofford Citation2003).
The complete mitochondrial genome of A. m. jemenitica was 16,623 bp long. It was AT-rich, consisting of 43.4% A, 41.5% T, 9.5% C, and 5.5% G. Of the 13 PCGs, nine were located on the light strand and four on the heavy strand. Four start codons were used: ATT (atp8, co2, nad1, nad4l, nad5, nad6), ATG (atp6, co3, cytb, nad4), ATA (co1, nad3), and ATC (nad2). All 13 PCGs used TAA as the stop codon. Atp8 and atp6 overlapped, sharing 19 bp. Of the 22 transfer RNAs (tRNAs), the shortest was tRNA-Gln with 63 bp and the longest at 80 bp was tRNA-Thr. The two rRNAs were both AT-rich and located on the heavy strand (16S: 1,358 bp, 84.6% AT; 12S: 788 bp, 81.5% AT).
Previous samples of A. m. jemenitica from Yemen had both O- and A-lineage haplotypes (Smith, unpublished, in Al-Ghamdi et al. Citation2013). Here, our sample was closest to A. m. lamarckii (p = 0.00381) and A. m. syriaca (p = 0.00586) (), suggesting it forms part of the Z subgroup of the A-lineage (Alburaki et al. Citation2011). Sequencing additional samples of A. m. jemenitica across its geographic distribution would be informative.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alburaki M, Moulin S, Legout H, Alburaki A, Garnery L. 2011. Mitochondrial structure of eastern honeybee populations from Syria, Lebanon and Iraq. Apidologie. 42(5):628–641.
- Al-Ghamdi AA, Nuru A, Khanbash MS, Smith DR. 2013. Geographical distribution and population variation of Apis mellifera jemenitica Ruttner. J Apic Res. 52(3):124–133.
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. [accessed 2018 Aug]. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bernt M, Pütz J, Florentz C, Donath A, Jühling F, Stadler PF, Externbrink F, Fritzsch G, Middendorf M. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boardman L, Eimanifar A, Kimball RT, Braun EL, Fuchs S, Grünewald B, Ellis JD. 2019. The complete mitochondrial genome of Apis mellifera unicolor (Insecta: Hymenoptera: Apidae), the Malagasy honey bee. Mitochondrial DNA Part B. 4(2):3286–3287.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Eimanifar A, T. Kimball R, L. Braun E, Fuchs S, Grünewald B, Ellis JD. 2017. The complete mitochondrial genome of Apis mellifera meda (Insecta: Hymenoptera: Apidae). Mitochondrial DNA Part B. 2(1):268–269.
- El-Niweiri MAA, Moritz R. 2008. Mitochondrial discrimination of honeybees (Apis mellifera) of Sudan. Apidologie. 39(5):566–573.
- Franck P, Garnery L, Loiseau A, Oldroyd BP, Hepburn HR, Solignac M, Cornuet J-M. 2001. Genetic diversity of the honeybee in Africa: microsatellite and mitochondrial data. Heredity (Edinb). 86(4):420–430.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Maddison WP, Maddison DR. 2018. Mesquite: a modular system for evolutionary analysis. Version. 3:5. [accessed 2018 Aug]. https://www.mesquiteproject.org/.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA; p. 1–8.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland (MA): Sinauer Associates.