Abstract
The mitochondrial genome of Apis mellifera ruttneri consisted of 13 protein-coding genes, two rRNAs, 22 tRNAs, an AT-rich control region, and was 16,577 bp long. The phylogenetic analyses suggested that A. m. ruttneri was closely related to two North African subspecies: A. m. sahariensis and A. m. intermissa.
The Maltese honey bee, Apis mellifera ruttneri (Sheppard et al.), is endemic to Malta. Despite this European location, analyses, including using mitochondrial DNA with EcoR1 restriction enzyme digestion, suggest that A. m. ruttneri forms part of the African (A) lineage - similar to A. m. sicula (Sheppard et al. Citation1997). Here, we present the mitochondrial genome of a worker A. m. ruttneri honey bee from San Julian’s, Malta (35°53 N, 14°26 E, collector: Dr. W.S. Sheppard, 1995, Voucher 2050 from Ruttner Bee Collection at the Bee Research Institute at Oberursel, Germany, GenBank: MN714162). Identity of the subspecies was confirmed morphometrically.
Following Eimanifar et al. (Citation2017), genomic DNA was extracted, quantified, and PE-150 bp sequenced on Illumina Hi-Seq 3000/4000 (San Diego, CA). The sequencing data were checked for quality using FastQC (Andrews Citation2010), and trimmed with Trimmomatic (Bolger et al. Citation2014). The paired R1 trimmomatic output was mapped to the mitogenome of A. m. sahariensis (MF351881) in Geneious Prime 2019.0.4 (Kearse et al. Citation2012) using the method from Boardman et al. (Citation2020). The assembled mitogenome was annotated in mitos2 (Bernt et al. Citation2013). The annotated mitogenome was manually adjusted to the A. m. capensis annotation (KX870183) in Geneious Prime. Protein-coding genes (PCGs) and ribosomal RNA (rRNA) sequences were manually aligned to sequences from other Apis species and Apis mellifera subspecies in Mesquite v3.5 (Maddison and Maddison Citation2018). Phylogenetic comparisons used RAxML 8.2.10 (Stamatakis Citation2014) with the GTRGAMMA model and 1000 bootstrap replicates (-f a option) on the CIPRES Science Gateway V.3.3 (Miller et al. Citation2010), and p-distances were generated with PAUP 4.0a (Swofford Citation2003).
The mitochondrial genome of A. m. ruttneri was 16,577 bp (43.2% A 9.6% C, 5.6% G, 41.5% T). Among the 13 protein-coding genes, nine were on the light strand (nad2, co1, co2, atp8, atp6, co3, nad3, nad6 and cytb) and four were on the heavy strand (nad1, nad4, nad4l, nad5). The two atp genes, atp8 and atp6, shared 19 nucleotides. The 13 PCGs used a variety of start codons: ATT (n = 6), ATG (n = 4), ATA (n = 2), and ATC (n = 1), while all PCGs ended with TAA. The 16S ribosomal RNA (rRNA) was 1,325 bp, with 84.1% AT, while the 12S rRNA was 785 bp with 81.2% AT. There were 22 transfer RNAs located in various positions on the mitogenome. They varied in size from threonine being the longest (80 bp) to serine and glutamine being the shortest (63 bp).
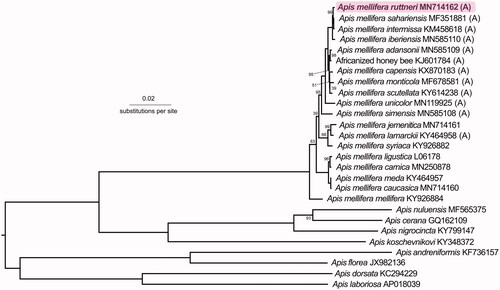
Phylogenetically, A. m. ruttneri was closest to other North African honey bees (). The subspecies with the lowest p-distances from A. m. ruttneri were A. m. sahariensis (p = 0.00114), A. m. intermissa (p = 0.00129), and A. m. iberiensis (p = 0.00221). This confirms previous studies suggesting that A. m. ruttneri is part of the A-lineage. Given their close geographic proximity, comparable size and color, and similar behavioral characteristics (Sheppard et al. 1997), future research comparing this mitogenome to that of the Sicilian honey bee A. m. sicula would be interesting.
Figure 1. Phylogenetic tree showing the relationship between Apis mellifera ruttneri (GenBank: MN714162) and 26 other Apis honey bees (GenBank accession numbers are listed after species names). Node labels indicate bootstrap values. Unlabeled lineages are 100%. The (A) indicates subspecies from the African A-lineage.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. [accessed 2019 Aug]. https//www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bernt M, Pütz J, Florentz C, Donath A, Jühling F, Stadler PF, Externbrink F, Fritzsch G, Middendorf M. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boardman L, Eimanifar A, Kimball RT, Braun EL, Fuchs S, Grunewald B, Ellis JD. 2020. The mitochondrial genome of Apis mellifera simensis (Hymenoptera: Apidae), an Ethiopian honey bee. Mitochondrial DNA Part B. 5(1):9–10.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Eimanifar A, T. Kimball R, L. Braun E, Fuchs S, Grünewald B, Ellis JD. 2017. The complete mitochondrial genome of Apis mellifera meda (Insecta: Hymenoptera: Apidae). Mitochondrial DNA Part B. 2(1):268–269.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Maddison WP, Maddison DR. 2018. Mesquite: a modular system for evolutionary analysis. Version. 3:5. [accessed 2018 Aug]. https//www.mesquiteproject.org/.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA; p. 1–8.
- Sheppard W, Arias M, Grech A, Meixner M. 1997. Apis mellifera ruttneri, a new honey bee subspecies from Malta. Apidologie. 28(5):287–293.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland (MA): Sinauer Associates.
