Abstract
Aquilegia barnebyi, belonging to the genus Aquilegia (Ranunculaceae), is a member of basal eudicot species. In this study, we obtained the complete chloroplast (cp) genome of A. barnebyi. The genome size is 161,954 bp with a GC content of 38.98%. A total of 113 unique genes including 79 protein-coding genes, 30 tRNA genes, four rRNA genes were annotated. The large single-copy region and small single-copy region contains 91,250 bp and 17,359 bp, respectively. The inverted repeat regions are 26,671 bp in length. The phyologenetic analysis indicated that A. barnebyi had a close relationship with A. coerulea. And four species in genus Aquilegia formed a monophyletic group with high support value. The availability of A. barnebyi cp genomic resources will greatly helpful for taxonomy, phylogeny and conservation genetic studies of basal eudicot specie.
The Aquilegia genus (Ranunculaceae) belongs to basal eudicot angiosperms which consists of approximately 75 species occurring in North America (22 spp), Asia (23 spp), and Europe (21 spp) (Nold Citation2003). These species have a number of characteristics that show their primitive evolution (Zahn et al. Citation2010). The investigation of the Aquilegia genus can serve as an evolutionary link between core eudicots and monocots. Furthermore, as a classic example of adaptive radiation, the Aquilegia genus has outstanding potential as a subject of evolutionary studies (Kramer and Hodges Citation2010). However, the little genetic and genomic information of the Aquilegia has hindered the study of the genus. In this study, we report the cp genome of A. barnebyi, thus providing useful information for taxonomy, evolutionary dynamics and conservation studies of the Aquilegia genus and basal eudicot angiosperms.
The leaves of A. barnebyi were collected from rifle falls of northwest Colorado, USA (39°40′35″N, 107°41′57″W), and the speciman was deposited at the herbarium of department of ecology, evolution and marine biology, university of California (speciman code Aquilegia_BA). The chloroplast reads were filtered from whole genome Illumina sequencing data of A. barnebyi which was downloaded from NCBI Short Read Archive (SRR7965809) (Filiault et al. Citation2018). We mapped all the sequencing reads to the A. rockii cp genome using bowtie2 (v2.3.4.3) (Langmead et al. Citation2009; Yu et al. Citation2019). The mapped chloroplast reads were assembled using Geneious v7.1.7 (Biomatters, New Zealand) with the A. rockii cp genome as reference. The complete cp genome was annotated using the program DOGMA (Wyman et al. Citation2004).Circular genome map was drawn using OGDRAW (Lohse et al. Citation2013).
The complete cp genome of A. barnebyi was composed of single circular double-stranded DNA molecules and was 161,954 bp in length. The data was deposited in GenBank with the accession number MN882557. As other taxa in the family of Ranunculaceae (Yu et al. Citation2019), the cp genome of A. barnebyi displayed the typical quadripartite structure, including a pair of inverted repeat regions (IR with 26,671 bp) divided by two single-copy regions (LSC 91,250 bp and SSC 17,359 bp). The overall GC content of the cp genome was 38.98%. There were a total of 113 unique genes, including 79 protein-coding genes, 30 tRNA genes and 4 rRNA genes, in A. barnebyi cp genome. Among these genes, nine protein-coding genes and six tRNA genes contained a single intron, and two protein-coding genes possessed two introns. The gene rps12 found to be trans-spliced; with the 5′-end exon located in the LSC region and two copies of 3′-end exon and intron in the IR regions. Moreover, the rpl32, infA and clpP were identified as pseudogenes because of the partial duplication.
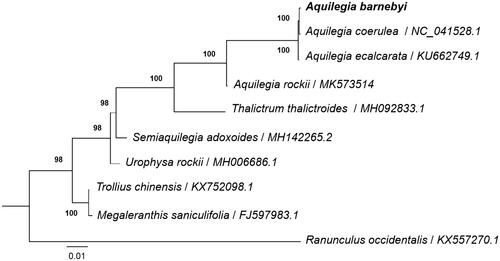
To understand the phylogenetic position of A. barnebyi within the genus Aquilegia and the family Ranunculaceae, we downloaded the complete cp genome of nine species in Ranunculaceae including three species in genus Aquilegia. The sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013), and RAxML (Stamatakis Citation2014) was used to construct a maximum likelihood tree with Ranunculus occidentalis as outgroup. All nodes in the complete plastome tree were strongly supported. The phylogenetic tree showed that four species in genus Aquilegia formed a monophyletic group with high support value. And A. barnebyi was closely related to A. coerulea and far away from A. rockii (). This published A. barnebyi cp genome will provide useful information for phyogenetic and evolutionary studies in the genus Aquilegia and Ranunculaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Filiault DL, Ballerini ES, Mandáková TM, Aköz G, Derieg NJ, Schmutz J, Jenkins J, Grimwood J, Shu S, Hayes RD, Hellsten U, et al. 2018. The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history. eLife. 7:e36426.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kramer EM, Hodges SA. 2010. Aquilegia as a model system for the evolution and ecology of petals. Phil Trans R Soc B. 365(1539):477–490.
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581.
- Nold R. 2003. Columbines. Aquilegia, paraquilegia and semiaquilegia. Cambridge (UK): Timber Press.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yu F, Zhao YC, Huang H. 2019. The complete chloroplast genome of aquilegiarockii, an endemic herb plant in Western China. Mitochondrial DNA B. 4(1):1737–1738.
- Zahn LM, Ma X, Altman NS, Zhang Q, Wall PK, Tian D, Gibas CJ, Gharaibeh R, Leebens-Mack JH, dePamphilis CW, et al. 2010. Comparative transcriptomics among floral organs of the basal eudicot Eschscholzia californica as reference for floral evolutionary developmental studies. Genome Biol. 11(10):101.