Abstract
In this study, a complete mitochondrial genome of a female Harpiocephalus harpia from Zhejiang Province, China, was sequenced using Illumina Hiseq. The genome is ∼16,400 bp in length, containing 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and a control region. Most of the genes were encoded on the H-strand, except for 8 tRNA and ND6 genes. Phylogenetic tree using maximum likelihood suggests H. harpia is sister taxon to Murina. The complete mitochondrial genome herein provides basic information for future taxonomic and phylogenetic studies.
Among all forest tube-nosed bats (Murininae), Harpiocephalus harpia (Chiroptera: Vespertilionidae) is the largest-sized and a phylogenetically basic species (Eger and Lim Citation2011; Simmons Citation2005; Soisook et al. Citation2013). Due to its strong sexual dimorphism, a long-term misclassification of H. harpia and H. mordax lasted until a comprehensive comparative analyses by Matveev (Citation2005), which evidenced H. mordax is synonym to H. harpia (Corbet and Hill Citation1992; Matveev Citation2005; Simmons Citation2005). Nowadays, the species is widely distributed in Southeast Asia, including India, Myanmar, Vietnam, Laos, Indonesia, Malaysia, and the Philippines (Smith and Xie Citation2008; Wilson et al. Citation2019). In China, it is recorded in Yunnan (Wang Citation2003), Guangdong (Zhou et al. Citation2014), Fujian (Jiang et al. Citation2015), Taiwan (Lin et al. Citation2006), Guangxi (Chen et al. Citation2015), Jiangxi (Chen et al. Citation2015), Hainan ( Hu et al. Citation2018), Hunan (Yu et al. Citation2017), Guizhou (Gong et al. Citation2018), Zhejiang (Yue, Hu et al. Citation2019), Hubei (Yue, Hu et al. Citation2019). It is believed that it suffers from habitat destruction due to agricultural expansion and deforestation, thus IUCN Red List and China Species Red List (Wang and Xie Citation2009; Wilson et al. Citation2019) list it as Least Concern and Vulnerable, respectively.
In this study, we determined characteristics of the complete mitochondrial genome (Genbank accession No. MN885881) basing upon an adult female H. harpia captured from Dongyang City, Zhejiang Province, China (29°10′ 37″ N 120°30′ 43″ E ). Presently, the specimen is stored in the Key Laboratory of Conservation and Application in Biodiversity of South China, Guangzhou University, Guangdong, China (Accession ID GZHU 17323). Complete genomic DNA was extracted from 20 mg liver tissue using MiniBEST Universal Genomic DNA Extraction Kit (TAKARA, Dalian) and was sequenced using Illumina Hiseq sequencing technology. Illumina PE library (460 bp library) was constructed and the whole genome of mitochondrial was scanned by bioinformatics analysis after quality control of the obtained sequencing data.
Published mitochondrial genome of M. ussuriensis (Yoon et al. Citation2013), M. leucogaster (Yoon and Park Citation2016), M. huttoni (Zhang et al. Citation2016), M. shuipuensis (Huang et al. Citation2019) and M. cyclotis (Yue, Huang et al. Citation2019) were used to annotate the mitochondrial genome of H. harpia. The complete mitochondrial genome is ∼16,400 bp in length, containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 1 control region. Among these 37 genes, most genes were encoded on the H-strand while ND6 (protein-coding gene) and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) were encoded on the L-strand. Among the 13 protein-coding genes, ATP8 and ATP6 were overlapped by 43 nucleotides, ND4L and ND4 were overlapped by 7 nucleotides. The initiation codon of most of the protein-coding genes was ATG, except for those of ND2 and ND5 (ATA), ND3 (ATC). Termination codon of Cyt b was AGA, seven protein-coding genes were TAA, while the rest were incomplete TA- (ND1 and ND3) or T-- (ND2, COX3, ND4). Incomplete termination codon will probably be completed by polyA of the 3′ end of mRNA after transcription (Ojala et al. Citation1981). The length of the 22 tRNA genes range from 62 to 75 bp and can be folded into typical cloverleaf secondary structure, with an exception of tRNA-Ser. The 12S rRNA gene was located between tRNA-Phe and tRNA-Val and 16S rRNA gene was located between tRNA-Val and tRNA-Leu. Lengths of them were 971 and 1560 bp, respectively. The D-loop region is located between tRNA-Pro and tRNA-Phe.
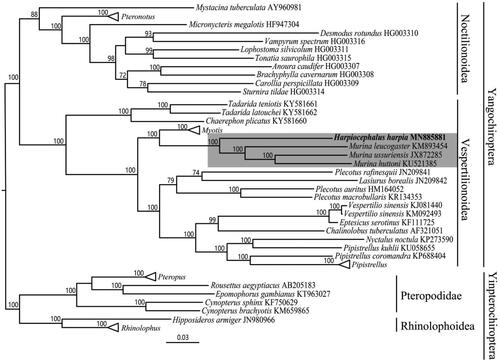
For phylogenetic inference, 64 released complete mitochondrial genome sequences of Chiropteran species were downloaded from the GenBank. Sequence matrix was aligned using the MUSCLE (Edgar Citation2004). PartitionFinder 1.1.1 (Lanfear et al. Citation2012) was used to select the best partitioning scheme and best-fit models of nucleotide evolution. We inferred maximum-likelihood (ML) tree using RAxML 8.2.4 (Stamatakis Citation2006, Citation2014) with 100 bootstraps setting. Our ML tree shows that H. harpia and Murina clade is a sister group and support a monophyly of Murininae (). The complete mitochondrial genome sequence of H. harpia herein could benefit the future taxonomic and phylogenetic studies.
Figure 1. ML phylogenetic trees of 65 chiropteran species based on complete mitochondrial genome. Numbers above the nodes indicate boot strap values. Branch length is based on ML trees. The shaded highlights are our sample and Murina. Hollow triangles represent clusters of multiple species of Pteropus, Rhinolophus, Pteronotus, Myotis and Pipistrellus genus including 8, 7, 7, 7 and 3 species respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen BC, Yu WH, Wu Y, Li F, Xu ZX, Zhang QP, Harada M, Motokawa M, Peng HY, Wang YY, et al. 2015. New record and sexual dimorphism of Harpiocephalus harpia in Guangxi and Jiangxi, China. Sichuan J Zool. 34(2):211–215.
- Corbet GB, Hill JE. 1992. The mammals of the Indomalayan region: a systematic review. Oxford: Oxford University Press.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Eger JL, Lim BK. 2011. Three New Species of Murina from Southern China (Chiroptera: Vespertilionidae). Acta Chiropterologica. 13(2):227–243.
- Gong LX, Gu H, Sun CN, Ma Q, Jiang TL, Feng J. 2018. Two new records of the Chiroptera in Guizhou Province (China) and their echolocation calls. Chin J Zool. 53(3):329–338.
- Hu YF, Li F, Wu Y, Li YC, Yu WH. 2018. New record of Harpiocephalus harpia in Hainan Province. J Zhejiang Forest Sci Technol. 38(3):85–88.
- Huang ZLY, Yue Y, Thapa S, Hu YF, Wu Y, Yu WH. 2019. Mitochondrial genome of Murina shuipuensis (Chiroptera: Vespertilionidae) from Shuifu Village, Guizhou, China (type locality). Mitochondr DNA B. 4(2):2588–2590.
- Jiang ZG, Ma Y, Wu Y, Wang YX, Zhou KY, Feng ZJ, Li LL. 2015. Chinàs mammal diversity and geographic distribution. Beijing: Sciences Press.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. Partition finder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Lin L-K, Harada M, Motokawa M, Lee L-L. 2006. Updating the occurrence of Harpiocephalus harpia (Chiroptera: Vespertilionidae) and its karyology in Taiwan. Mammalia. 70(1–2):170–172.
- Matveev VA. 2005. Checklist of Cambodian bats (Chiroptera), with new records and remarks on taxonomy. RusJTheriol. 4(1):43–62.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474.
- Simmons NB. 2005. Order Chiroptera. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Baltimore: The Johns Hopkins University Press.
- Smith AT, Xie Y. 2008. A guide to the mammals of China. Princeton: Princeton University Press.
- Soisook P, Karapan S, Satasook C, Bates P. 2013. A new species of Murina (Mammalia: Chiroptera: Vespertilionidae) from peninsular Thailand. Zootaxa. 3746(4):567–579.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang YX. 2003. A complete checklist of mammal species and subspecies in China: a taxonomic and geographic reference. Beijing: China Forestry Publishing House.
- Wang S, Xie Y. 2009. China Species Red List. Vol. II Vertebrates. Beijing: Higher Education Press.
- Wilson DE, Mittermeier RA, Edicions L. 2019. Handbook of the mammals of the world. In: Bats. Vol. 9. Barcelona, ES: Lynx Edicions.
- Yoon KB, Kim HR, Kim JY, Jeon SH, Park YC. 2013. The complete mitochondrial genome of the Ussurian tube-nosed bat Murina ussuriensis (Chiroptera: Vespertilionidae) in Korea. Mitochondr DNA. 24(4):397–399.
- Yoon GB, Park YC. 2016. The complete mitogenome of the Korean greater tube-nosed bat, Murina leucogaster (Chiroptera: Vespertilionidae). Mitochondr DNA A. 27(3):2145–2146.
- Yu WH, Hu YF, Guo WJ, Li F, Wang XY, Li YC, Wu Y. 2017. New discovery of Harpiocephalus harpia in Hunan Province and its potential distribution area in China. J Guangzhou Univ Nat Sci Ed. 16(3):16–20.
- Yue Y, Hu YF, Lei BY, Wu Y, Wu H, Liu BQ, Yu WH. 2019. Sexual dimorphism in Harpiocephalus harpia and its new records from Hubei and Zhejiang, China. Acta Theriol Sin. 39(2):142–154.
- Yue Y, Huang ZLY, Li F, Thapa S, Hu YF, Wu Y, Yu W. 2019. The complete mitochondrial genome of the tube-nosed bat Murina cyclotis (Chiroptera: Vespertilionidae) in China. Mitochondr DNA B. 4(2):2248–2250.
- Zhang QP, Cong HY, Yu WH, Kong LM, Wang YY, Li YC, Wu Y. 2016. The mitochondrial genome of Murina huttoni rubella (Chiroptera: Vespertilionidae) from China. Mitochondr DNA B. 1(1):438–440.
- Zhou Q, Xu ZX, Yu WH, Li F, Chen BC, Gong YN, Harada M, Motokawa M, Li YC, Wu Y. 2014. The occurrence of bat Harpiocephalus harpia from Nanlin, Guangdong and its karyotypes, echolocation calls. Chin J Zool. 49(1):41–45.
