Abstract
We assembled and characterized the complete chloroplast genome sequence of Silvianthus bracteatus to investigate its phylogenetic position. With a total length of 155,125 bp, the plastome comprised of a large single-copy (LSC) region of 86,054 bp, a small single-copy (SSC) region of 17,625 bp, and two inverted repeat (IR) regions of 25,723 bp. The overall percentage of GC content was 37.9. The new sequence comprised of total 136 genes, including 88 protein-coding genes, 8 ribosomal RNA genes, and 40 tRNA genes. In these genes, eight genes contained one intron and two genes contained two introns. Phylogenetic analysis showed that S. bracteatus was close to the Oleaceae.
The Carlemanniaceae is a family of shrub to herbaceous perennial flowering plants, including only two genera (Carlemannia and Silvianthus) and four species. Taxonomists had put the two genera into the Caprifoliaceae or the Rubiaceae (Chen and Anthony Citation2011). The Angiosperm Phylogeny Group placed the group in the Lamiales and as a sister group to the Oleaceae and the systematic position of Carlemanniaceae is still largely unknown (Stevens Citation2017). Here, we report and characterize the first complete plastome of Silvianthus bracteatus, an endemic shrub in the northern Indochina peninsula to investigate its phylogenetic position (GenBank accession number: MN908147).
The leaves of S. bracteatus was sampled from Xishuangbanna, China (21.9658°N, 101.2181°E) and we deposited the voucher specimen at the Herbarium of Guizhou Normal University (Accession number: Zhu201905004). Total genomic DNA was extracted with the Qiagen DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, USA). The genomic paired-end (PE150) sequencing was performed on an Illumina Hiseq 2500 instrument. The cp genome was assembled using the program GetOrganelle v1.6.2e (Jin et al. Citation2018). Annotation was performed using PGA (Qu et al. Citation2019) and manually corrected.
A typical quadripartite structure as most angiosperms was displayed by the plastome of S. bracteatus, containing two inverted repeat regions of 25,723 bp, a large single-copy region of 86,054 bp, and a small single-copy region of 17,625 bp. The new sequence comprised total 136 genes, including 88 protein-coding genes, 8 ribosomal RNA genes, and 40 tRNA genes. In these genes, eight protein-coding genes (atpF, ndhA, ndhB, petD, rpl16, rpl2, rps12, rps16) contained one intron and two genes (clpP and ycf3) contained two introns. The overall percentage of GC content was 37.9 and the corresponding value of the LSC, SSC, and IR region were 35.9, 32.4, and 43.1, respectively.
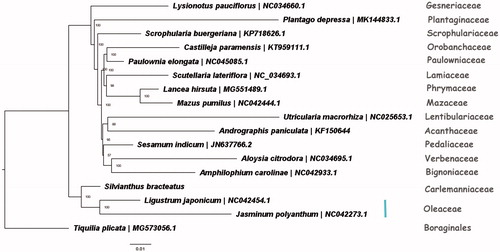
Fifteen complete chloroplast genome sequences of Lamiales and one complete chloroplast genome sequences of Boraginales were downloaded from NCBI to further investigate the phylogenetic position of S. bracteatus. We constructed a maximum likelihood tree using RAxML (Stamatakis Citation2014) after the sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013). Our results showed that S. bracteatus was close to the Oleaceae (). This published S. bracteatus chloroplast genome will provide useful information for phylogenetic and evolutionary studies in Carlemanniaceae and Lamiales.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen T, Anthony RB. 2011. Flora of China, Volume 19: Carlemanniaceae. In: Wu Z-Y, Raven PH, Hong D-Y, editors. Beijing: Science Press; St. Louis, Missouri: Missouri Botanical Garden Press.
- Jin J-J, Yu W-B, Yang J-B, Song Y, Yi T-S, Li D-Z. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. 256479. doi:10.1101/256479.
- Stevens PF. 2017. Angiosperm Phylogeny Website. Version 14. Published on the Internet. [accessed 2012 Jan 1]. http://www.mobot.org/MOBOT/Research/APweb/.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment soft-ware version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.