Abstract
Complete mitogenome of the leafhopper species Reticuluma hamata are sequenced and assembled and annotated, the mitogenome is 15,190 bp, the base composition is 43.5% for A, 12.4% for C, 9.4% for G, and 34.7% for T. The mitogenome includes 13 protein-coding genes, 22 transfer RNA, and 2 ribosomal RNA genes. Within phylogenetic tree, all Deltocephalinae members were clustered into a clade with strongly supported. This study enriches the mitogenomes of the leafhopper.
Keywords:
Reticuluma belonging to the tribe Penthimiini of the subfamily Deltocephalinae (Hemiptera: Cicadellidae), which contained 8 species until now, and exclusively distributed in China (Wang and Zhang Citation2019). Currently, the mitogenome of the tribe Penthimiini has been not recorded. In this study, the complete mitogenome sequences of R. hamata are the first time to determine, we hope that will apply to the studies of the identification and evolution of Penthimiini leafhoppers.
The genome DNA was extracted from adult of R. hamata which were collected from Yunnan Province, China (E 101°30′9″, N 23°57′50″), in June 2019. All the specimens were preserved in absolute alcohol and deposited in the Institute of Entomology of Guizhou University, Guiyang, China (GUGC-HCD-00113). The complete mitogenome of R. hamata forms a typical closed loop with a full length of 15,190 bp. This resource of sequence and annotation was submitted to NCBI (GenBank accession number MN922303). The composition of the mitogenome is as follows: 13 PCGs, 22 tRNA genes, 2 rRNA genes and a long non-coding region, A = 43.5%, C = 12.4%, G = 9.4%, T = 34.7%, with a total A + T content of 78.2% and is within the range reported within Hemipteran mitogenomes (68.86–86.33; Wang et al. Citation2015). Five PCGs (ND3, ND4, ND4L, ND6, ND1) started with ATT, four PCGs (ND2, COII, ATP6, CytB) started with ATA, two PCGs (COI, ATP8) started with ATC, two PCGs (COIII, ND5) started with ATG. COI and ND3 stopped with TAG, the remaining eleven PCGs stopped with TAA. All tRNA genes are identified by ARWEN version 1.2 software (Laslett and Canbäck Citation2008). The lengths of 16S rRNA gene and 12S rRNA gene are 1,190 bp and 723 bp, respectively. They are located between trnL2(CUN) and trnV, trnV and Control region, respectively. The control region is 904 bp long, and is located between 12S rRNA and trnI.
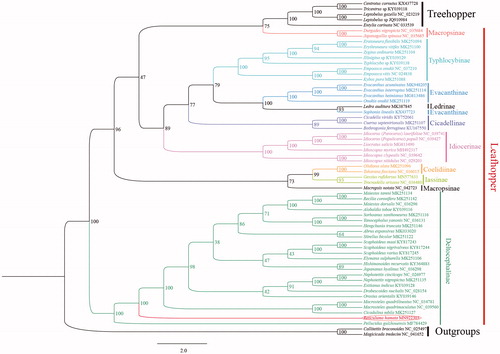
The phylogenetic relationships of R. hamata were reconstructed with IQ-TREE using an ultrafast bootstrap approximation approach with 10,000 replicates based on concatenated the nucleotides of the 13 PCGs. Each PCG sequences were aligned using the MAFFT algorithm in TranslatorX and aligned sequences were eliminated using Gblocks 9.1 b (Abascal et al. Citation2010). In the phylogenetic tree, all Deltocephalinae members were clustered into a clade with strongly supported (bootstrap support value = 100). The result confirms that R. hamata is part of Deltocephalinae (). On the other hand, this study can provide molecular data for further phylogenetic and evolutionary analysis, we hope this data can be useful for further study.
Disclosure statement
The authors declare no potential conflict of interest was reported.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38(suppl_2):W7–13.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Wang Y, Chen J, Jiang LY, Qiao GX. 2015. Hemipteran mitochondrial genomes: features, structures and implications for phylogeny. IJMS. 16(12):12382–12404.
- Wang DM, Zhang YL. 2019. Two new species of the genus Reticuluma Cheng & Li (Hemiptera: Cicadellidae: Deltocephalinae: Penthimiini) from China. Zootaxa. 4668(2):289–295.