Abstract
The complete mitochondrial genome sequence of Coronocyclus labratus was sequenced in the present study. It was determined to be 13,856 bp in length, containing 12 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 2 non-coding regions. The nucleotide sequence data of 12 protein-coding genes of C. labratus and other 16 Strongylidae species were used for phylogenetic analyses. Coronocyclu labratus formed a monophyletic cluster with the remaining Strongylidae species. Coronocyclus genus was present in the same clade with high statistical support. The mitogenome sequences will facilitate taxonomy as well as systematic studies of Cyathostominae nematodes.
Cyathostominae and Strongylinae are the two subfamilies of Strongylidae nematodes, which inhabit in the large intestine with a high prevalence in Equus species. They cause series clinical symptoms such as anemia, weight loss, and even death (Lichtenfels et al. Citation2002). Coronocyclus labratu, which belongs to subfamily Cyathostominae, is a significant Strongylidae nematode. In the present study, we assembled and characterized the complete mitochondrial genome of C. labratus from horse and to reconstruct the phylogenetic relationships of Strongylidae nematodes.
Specimen (Specimen Voucher: Biological Specimen Museum of Henan Normal University #YJ0096) were isolated from intestine of infected donkey in Yanjin, Henan Province, China (114°19′E, 33°14′N). Genomic DNA of the specimen was extracted using standard phenol/chloroform methods (Sambrook et al. Citation1989). Illumina HiSeq 4000 platform were used to amplify the entire mitochondrial genome sequence (GenBank accession number: MN832736). The complete mitogenome sequence of C. labratus was assembled with SOAPdenovo v2.04 (Luo et al. Citation2012) and MITObim v1.6 (Christoph et al. Citation2013). The complete mitogenome was annotated using the DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Lowe and Eddy Citation1997).
The complete mitochondrial genome of C. labratus is 13,856 bp in length. The complete mitochondrial genome has 12 protein-coding genes, 22 transfer RNA genes (tRNA), and 2 ribosomal RNA (rRNA, rrnL, and rrnS) genes, all genes are encoded by the same strand. Two kinds of start codons (ATT and TTG) and two kinds of termination codons (TAA and T) were used in the 12 PCGs, respectively. The studied genome has a high T content (45.62%) and a low C content (6.96%), resulting in a very strong A + T bias (76.91%) and in particular, the AT-rich region (85.63%). The size of the 22 tRNAs ranging from 52 to 63 bp. The rrnL (975 bp) is located between trnH and nad3, whereas the rrnS (701 bp) is located between trnE and trnS(UCN).
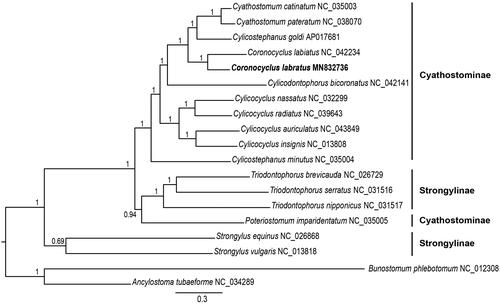
Phylogenetic analyses were conducted using the Bayesian inference (BI), implemented in MrBayes version 3.1.2 (Ronquist and Huelsenbeck Citation2003). Mitogenome sequences of 16 other Strongylidae species were retrieved from GenBank. Analyses were performed using 12 protein-coding genes, using Bunostomum phlebotomum and Ancylostoma tubaeforme as outgroup. Phylogenetic analysis showed that Coronocyclus labratus formed a monophyletic cluster with the remaining Strongylidae species. Coronocyclus genus was present in the same clade with high statistical support (). Strongylidae nematodes grouped into two major clades: Strongylus species and others. Species of genus Triodontophorus (T. brevicauda, T. serratus, and T. nipponicus) clustered together with species in subfamily Cyathostominae, although they belonged to subfamily Strongylinae. The dendrogram topology is highly congruent with previous studies (Gao. et al. Citation2017; Li et al. Citation2019). The mitogenome sequences will facilitate taxonomy as well as systematic studies of Cyathostominae nematodes in the future.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Christoph H, Lutz B, Bastien C. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Gao Y, Zhang Y, Yang X, Qiu JH, Duan H, Xu WW, Chang QC, Wang CR. 2017. Mitochondrial DNA evidence supports the hypothesis that Triodontophorus species belong to Cyathostominae. Front Microbiol. 8:1444.
- Li Q, Gao Y, Wang XX, Li Y, Gao JF, Wang CR. 2019. The complete mitochondrial genome of Cylicocylus ashworthi (Rhabditida: Cyathostominae). Mitochondr DNA B. 4(1):1225–1226.
- Lichtenfels JR, Gibbons LM, Krecek RC. 2002. Recommended terminology and advances in the systematics of the Cyathostominea (Nematoda: Strongyloidea) of horses. Vet Parasitol. 107(4):337–342.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 5(5):955–964.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18.
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Sambrook T, Fritsch E, Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.