Abstract
The complete mitochondrial genomes of two toxic flatworm species of the genus Planocera were determined. The total length of mitochondrial DNA (mtDNA) was 15,657 bp in Planocera multitentaculata and 15,486 bp in Planocera reticulata and included 12 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and 4 non-coding regions. The mitochondrial gene arrangement of these planocerid species was identical to that of previously described acotylean species, Hoploplana elisabelloi, and also to those of other polyclads. Maximum likelihood analysis against 14 Platyhelminthes showed that a tree was robustly constructed using 12 protein-coding genes than COI gene.
Polyclad flatworms are found in coastal waters of the world. In these, several species in the Pacific Ocean are known as tetrodotoxin (TTX)-bearing flatworms, such as Planocera multitentaculata and Planocera reticulata, which are sympatric sister species in the Japanese waters (Miyazawa et al. Citation1986; Tanu et al. Citation2004; Yamada et al. Citation2017; Itoi et al. Citation2018). Here, we describe complete mitochondrial genome sequences of Planocera multitentaculata and Planocera reticulata.
Planocerid flatworms were collected at an intertidal zone in Hayama, Kanagawa, Japan (35°25′ N, 139°57′ E) in May 2017. Planocera multitentaculata (specimen num. UMUT-RW33197) and P. reticulata (specimen num. UMUT-RW33198) were preserved in 99% ethanol and stored in The University Museum, the University of Tokyo. Total genomic DNA was extracted from a tissue using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) and libraries were prepared using TruSeq Nano Library Prep kit (Illumina, San Diego, CA, USA). Next-generation sequencing analysis was performed using HiSeqX (Illumina) and sequences were assembled by CLC Genomics Workbench ver. 8.0.1 (Qiagen). Gene annotation including rRNA estimation and tRNA prediction was done using Geneious ver. 11.1.5 (Tomy Digital Biology, Tokyo, Japan) and Mitos WebServer (Bernt et al. Citation2013). The phylogenetic tree was constructed by means of the maximum likelihood method using MEGA X (Kumar et al. Citation2018).
A total mtDNA length of P. multitentaculata (LC503532) was 15,657 bp composed by 30.8% adenine, 12.4% cytosine, 40.0% thymine, and 16.9% guanine, whereas that of P. reticulata (LC503531) was 15,486 bp composed of 31.1% adenine, 11.2% cytosine, 41.1% thymine, and 16.7% guanine. The size of these mitochondrial genomes of both planocerids were smaller than those of other metazoan groups and all mitochondrial structural genes (2 rRNA genes, 22 tRNA genes, 12 protein-coding genes lacking ATPase 8) are encoded in the same strand as in the case of other flatworms (Gissi et al. Citation2008; Wey-Fabrizius et al. Citation2013; Golombek et al. Citation2015; Aguado et al. Citation2016). The ATG, GTG, TTG, and ATA start codons were used by P. multitentaculata, while the ATG and TTG counterparts were done by P. reticulata (Table S1)Footnote1. The TAA stop codon was largely used in protein-coding genes of both flatworms (Table S1). Control region could not be identified as in the case of other Platyhelminthes (Huyse et al. Citation2007), although four non-coding regions were detected (Aguado et al. Citation2016; Table S1). The identities of mtDNA sequences from two planocerids showed 95.8–97.5%, 86.6–100%, and 90.9–94.8% in rRNA, tRNA, and protein-coding genes, respectively (Table S1). The mitochondrial gene arrangements of two planocerids were identical to those in other flatworms (Aguado et al. Citation2016).
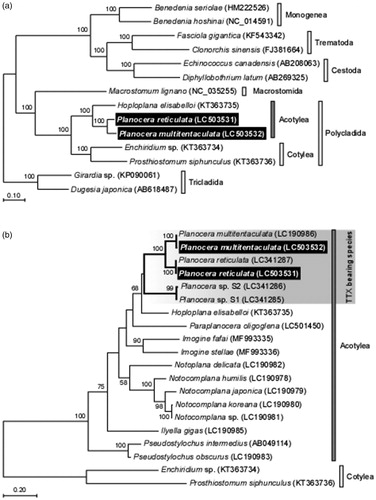
Maximum likelihood analysis based on mitochondrial genome sequences (12 protein-coding genes) from 14 Platyhelminthes support the monophyly of Polycladida including Acotylea and Cotylea () and that based on partial COI gene sequences revealed that the TTX-bearing flatworms formed a cluster containing the planocerid sequences (). Thus, the characterization of the planocerid mtDNA proved to be useful for understanding the phylogeny and taxonomy of flatworms and also for the development of molecular tools needed for investigating the ecology of these flatworms.
Figure 1. Phylogenetic relationship of two planocerid flatworms related Platyhelminthes species inferred from the 12 protein-coding genes on mitochondrial genome sequence (a) and COI gene (b). The phylogenetic tree was generated by maximum likelihood analysis under the General Time Reversible model. Numbers at branches denote the bootstrap percentages from 1000 replicates. Only bootstrap values exceeding 50% are presented. LC503531 and LC503532 in parentheses indicate the accession numbers deposited in DDBJ/EMBL/GenBank databases in this study and the accession numbers for reference sequences are shown in parentheses. Data used for the analysis were obtained from Planocera reticulata (LC341287), Planocera multitentaculata (LC190986), Hoploplana elisabelloi (KT363735), Planocera sp. S1 (LC341285), Planocera sp. S2 (LC341286), Paraplanocera oligoglena (LC501450), Imogine fafai (MF993335), Imogine stellae (MF993336), Ilyella gigas (LC190985), Notoplana delicata (LC190982), Notocomplana humilis (LC190978), Notocomplana japonica (LC190979), Notocomplana sp. (LC190981), Notocomplana koreana (LC190980), Pseudostylochus obscurus (LC190983), Pseudostylochus intermedius (AB049114), Enchiridium sp. (KT363734), Prosthiostomum siphunculus (KT363736), Macrostomum lignano (NC_035255), Girardia sp. (KP090061), Dugesia japonica (AB184487), Benedenia seriolae (HM222526), Benedenia hoshinai (NC_014591), Fasciola gigantica (KF543342), Clonorchis sinensis (FJ381664), Echinococcus canadensis (AB208063) and Diphyllobothrium latum (AB269325). The scale indicates number of nucleotide substitutions per site.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes
1 Table S1 is available on at FigShare: DOI 10.6084/m9.figshare.11709648.
References
- Aguado MT, Grande C, Gerth M, Bleidorn C, Carolina Noreña C. 2016. Characterization of the complete mitochondrial genomes from Polycladida (Platyhelminthes) using next-generation sequencing. Gene. 575(2):199–205.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Gissi C, Iannelli F, Pesole G. 2008. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 101(4):301–320.
- Golombek A, Tobergte S, Struck TH. 2015. Elucidating the phylogenetic position of Gnathostomulida and first mitochondrial genomes of Gnathostomulida, Gastrotricha and Polycladida (Platyhelminthes). Mol Phylogenet Evol. 86:49–63.
- Huyse T, Plaisance L, Webster BL, Mo TA, Bakke TA, Bachmann L, Littlewood DT. 2007. The mitochondrial genome of Gyrodactylus salaris (Platyhelminthes: Monogenea), a pathogen of Atlantic salmon (Salmo salar). Parasitology. 134(Pt 5):739–747.
- Itoi S, Ueda H, Yamada R, Takei M, Sato T, Oshikiri S, Wajima Y, Ogata R, Oyama H, Shitto T, et al. 2018. Including planocerid flatworms in the diet effectively toxifies the pufferfish, Takifugu niphobles. Sci Rep. 8(1):12302.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Miyazawa K, Jeon JK, Maruyama J, Noguchi T, Ito K, Hashimoto K. 1986. Occurrence of tetrodotoxin in the flatworm Planocera multitentaculata (Platyhelminthes). Toxicon. 24(7):645–650.
- Tanu MB, Mahmud Y, Arakawa O, Takatani T, Kajihara H, Kawatsu K, Hamano Y, Asakawa M, Miyazawa K, Noguchi T. 2004. Immunoenzymatic visualization of tetrodotoxin (TTX) in Cephalothrix species (Nemertea: Anopla: Palaeonemertea: Cephalotrichidae) and Planocera reticulata (Platyhelminthes: Turbellaria: Polycladida: Planoceridae). Toxicon. 41:515–520.
- Wey-Fabrizius AR, Podsiadlowski L, Herlyn H, Hankeln T. 2013. Platyzoan mitochondrial genomes. Mol Phylogenet Evol. 69(2):365–375.
- Yamada R, Tsunashima T, Takei M, Sato T, Wajima Y, Kawase M, Oshikiri S, Kajitani Y, Kosoba K, Ueda H, et al. 2017. Seasonal changes in the tetrodotoxin content of the flatworm Planocera multitentaculata. Mar Drugs. 15(3):56.
