Abstract
Pseudocrossocheilus tridentris is an endemic fish species in the upper Pearl River with limited published genetic information. In the current study, the complete mitochondrial genome of P. tridentris was determined using next generation sequencing, which was 16,606 base pairs (bp) in length and 91.65% identical to its closest relative with a genome of P. bamaensis. Phylogenetic analysis showed that P. tridentris had a close relationship with its congener P. bamaensis.
Introduction
Pseudocrossocheilus tridentris, belonging to the subfamily Labeoninae within Cyprinidae, is an endemic species exclusively distributed in the Nanpanjiang river, a mainstem in the upper Pearl River (Cui and Chu Citation1986). This species was initially assigned into genus Sinocrossocheilus till several researchers consistently considered it should be belonged to genus Pseudocrossocheilus (Cui and Chu Citation1986; Yuan et al. Citation2008). Given that narrow distribution, small population and easily influenced by environmental disturbances, this species is in urgent need of protecting and managing. To date, however, little reported genetic information of P. tridentris and no published complete mitochondrial genome have imposed restrictions on management and conservation of this endemic species.
Pseudocrossocheilus tridentris was collected during March in 2019 from the Maling River, a tributary within the upper Pearl River drainage. A bit of fin tissues were preserved in 99% ethanol for further DNA extraction. The specimen (specimen accession number: MLPT-20190312001) and tissues were stored in the museum of the Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences. Total genomic DNA was extracted from fin tissues using a Genomic DNA Isolation Kit (QiaGene, Hilden, Germany). We performed on the Illumina MiSeq platform to sequence the complete mitochondrial genome (Illumina Inc, San Diego, CA, USA). The Illumina raw sequence reads were assembled into contigs using SPAdes 3.9.0 (Bankevich et al. Citation2012) and then we assembled the contigs into the complete mitochondrial genomes implemented in SOAPdenovo (Luo et al. Citation2012).
The complete mitogenome sequence of P. tridentris (GenBank: MN794204) is 16,606 base pairs (bp) in length and contains the standard structures reported in most of teleost mitogenome including 13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes. The complete mitogenome of P. tridentris and 38 additional mitogenomes including 33 Labeoninae species and five outgroups were aligned using MUSCLE (Edgar Citation2004). All 13 protein-coding genes were subjected to concatenated into a single sequence for phylogenetic analysis using Bayesian inference technique. Nucleotide substitution model (GTR + I+G) were chosen based on the Akaike information criterion (AIC) in MRMODELTEST version 2.3 (Nylander Citation2004). The Bayesian analyses were performed in MrBayes 3.1.2 (Ronquist and Huelsenbeck Citation2003) and each run was conducted for 20 million generations and sampled every 1000 generations, with the first 25% discarded as burn-in.
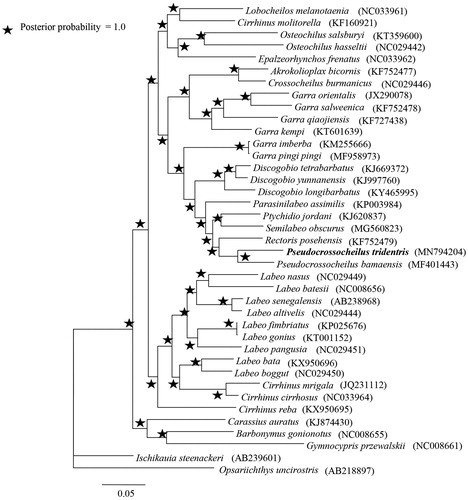
Bayesian trees revealed four well supported major clades with high supported values (Clade I, II, III, and IV; ). Pseudocrossocheilus tridentris is highly supported (100%) as the sister species of its congener P. bamaensis (). The P. tridentris mitogenome is 8.35% divergent from P. bamaensis based on Kimura 2-parameter model (Kimura Citation1980). In addition, we found that some genera (i.e. Cirrhinus, Garra and Labeo) did not display monophyly, indicating many taxonomic problems in this subfamily. More work on morphological traits and phylogenetic inferences should be conducted to understand the relationships within the Labeoninae in future.
Ethical approval
Experiments were performed in accordance with the recommendations of the Ethics Committee of Huaiyin Institute of Technology. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Acknowledgments
We are grateful to Mingsheng Wang, Zhijun Jin, Jun Wang, and Qiusheng Zhao for providing assistance in field sampling.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich A A, Dvorkin M, Kulikov A S, Lesin V M, Nikolenko S I, Pham S, Prjibelski A D, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Cui G, Chu X. 1986. New material for the Chinese Cyprinid genus Sinocrossocheilus. Acta Zootaxonomica Sinica. 11:425–428.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16(2):111–120.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18.
- Nylander J. 2004. MrModeltest v2. Evolutionary Biology Centre, Uppsala University.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Yuan LY, Zhang E, Huang YF. 2008. Revision of the labeonine genus Sinocrossocheilus (Teleostei: Cyprinidae) from south China. Zootaxa. 1809(1):36–48.