Abstract
In this study, the complete mitochondrial genome of Hydrelia parvulata (Staudinger, 1897) was firstly sequenced by high-throughput sequencing. As a circular DNA molecule, it is 15,407 bp in length (GenBank accession number: MN962739), which consists of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and an AT-rich region. The nucleotide composition is A (39.8%), C (11.6%), G (8.2%), and T (40.4%). Based on the sequences of complete mitogenome from 10 geometrid species as ingroups and 3 bombycid species as outgroups, the phylogenetic trees were constructed using maximum likelihood (ML) method. The results showed that H. parvulata has a close evolutionary relationship with Operophtera brumata in the subfamily Larentiinae, which is a clade strongly supported by the bootstrap value of 100% and a sister-group with the subfamily Ennominae.
Hydrelia parvulata (Staudinger, 1897) that is distributed in Russia, Korean Peninsula and Northern China belongs to Larentiinae in the family Geometridae (Lepidoptera: Geometriodea) (Xue and Scoble Citation2002; Tóth et al. Citation2018). In this study, the adults of H. parvulata were collected on July 25, 2019 in Huangbaiyuan Town (33.8152°N, 107.6633°E, Taibai County, Baoji City, Shaanxi Province) and its complete mitochondrial genome was reported for the first time. All specimens and the genomic DNA were deposited at the Insect Collection of Hunan Agricultural University, Changsha City, Hunan Province, China. We have obtained the complete mitogenome of H. parvulata and constructed a phylogenetic tree for understanding its phylogenetic position in the family Geometridae, and then analyzed the relationships with other closely related groups.
The complete genome DNA was extracted from the adult (collection number: HAUHL042201) using TaKaRa MiniBEST Universal Genomic DNA Extraction Kit Ver.5.0 (Shiga Prefecture, Kusatsu City, Japan) and was sequenced by using high-throughput sequencing in Novogene (Chen et al. Citation2019). The assembly of sequence used IDBA in server. The gene annotation was accomplished with ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) and MITOS web server (http://mitos.bioinf.uniLeipzig.de/index.py) (Bernt et al. Citation2013). To construct the phylogenetic tree, the complete mitogenomes of 10 ennomin and larentiin speciesas ingroups and 3 bombycid species (Wang et al. Citation2019) as outgroups were used. The maximum likelihood (ML) method was performed by MEGA 10 and the bootstrap analysis was set as 1000 pseudo-replicates.
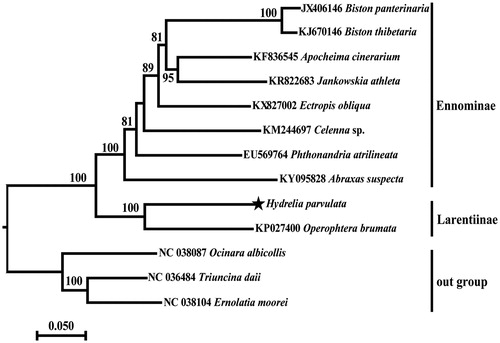
The complete mitogenome of H. parvulata is a circular DNA molecule of 15,407 bp in length (GenBank accession number: MN962739) and consists of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and an AT-rich region, in which 24 genes are transcribed on the J strand and the remaining 13 are transcribed on the N strand. The nucleotide composition is A (39.8%), C (11.6%), G (8.2%), T (40.4%), and the AT nucleotide content is 80.2%. There were 135 bp intergenic nucleotides that were dispersed in between 14 pairs of neighboring genes with their length varying from 1 to 25 bp. There were 17 bp overlapping nucleotides that were dispersed in between 3 pairs of neighboring genes with their length in 1, 7 and 8 bp. The length of the A + T rich region that was located between 12SrRNA and trnM was 412 bp. The phylogenetic tree based on sequences of the complete mitogenome was shown in . The relationship between the subfamilies Ennominae and Larentiinae, both of them are monophyletic groups, as the sister group is strongly supported by the bootstrap value of 100%. The species H. parvulata has a close evolutionary relationship with Operophtera brumata in the subfamily Larentiinae that is a clade strongly supported by the bootstrap value of 100%.
Figure 1. The evolutionary relationships of Hydrelia parvulata shown by the maximum-likelihood tree based on the complete mitogenomes of 13 lepidopteran moths. All the species accession numbers in this study are listed as below: Abraxas suspecta (KY095828), Apocheima cinerarium (KF836545), Biston panterinaria (JX406146), Biston thibetaria (KJ670146), Celenna sp. (KM244697), Ectropis obliqua (KX827002), Ernolatia moorei (NC 038104), Hydrelia parvulata (MN962739), Jankowskia athleta (KR822683), Operophtera brumata (KP027400), Ocinara albicollis (NC 038087), Phthonandria atrilineata (EU569764), Triuncina daii (NC 036484).
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication. The authors also are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Juling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadier PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen L, Liao CQ, Wang X, Tang SX. 2019. The complete mitochondrial genome of Gibbovalva kobusi (Lepidoptera: Gracillariidae). Mitochondrial DNA Part B. 4(2):2769–2770.
- Tóth B, Gergely K, Zsolt B. 2018. Data of Geometridae (Lepidoptera) from the Korean Peninsula in the collections of the Hungarian Natural History Museum – subfamily Larentiinae. Folia Ent Hung. 79:127–161.
- Wang X, Chen ZM, Gu XS, Wang M, Huang GH, Zwick A. 2019. Phylogenetic relationships among bombycidae s.l. (lepidoptera) based on analyses of complete mitochondrial genomes. Syst Entomol. 44(3):490–498.
- Xue DY, Scoble MJ. 2002. A review of the genera associated with the tribe Asthenini (Lepidoptera: Geometridae: Larentiinae). Bull Nat Hist Mus Entomol Ser. 71(1):77–133.
