Abstract
Pimpinella smithii Wolff is endemic in China. In this study, the complete chloroplast genome of P. smithii was determined. The total genome size was 147,130 bp in length. It was found to possess the quadripartite structure, containing a pair of inverted repeats (IRs) of 18,264 bp, which were separated by large single copy (LSC) and small single copy (SSC) of 93,158 bp and 17,444 bp, respectively. A total of 129 genes were identified, including 85 protein-coding genes, 36 tRNA genes, and eight rRNA genes. The maximum-likelihood phylogenetic analysis indicated that P. smithii has close relationship with genus Angelica.
Pimpinella smithii Wolff (Apiaceae) is an endemic plant and naturally distributed in China, growing in forest margins and grasslands at an elevation of 1400–3600 m (Pu and Watson Citation2005). Previous molecular phylogenetic analysis (Wang et al. Citation2014) showed that P. smithii was not closely allied with Pimpinella ‘core group’ in tribe Pimpinelleae, falling within the tribe Selineae. The complete chloroplast genome of P. smithii was assembled herein to further explore the reasonable relationship between this species and other allies.
The fresh leaves of P. smithii were collected from Songpan County, Sichuan Province, China (32°45′46″N, 103°52′38″E), and voucher specimens (W19080802) were deposited at HYNU (Hengyang Normal University Herbarium, Hengyang City, Hunan Province, China). Morphological characters were measured using Karyotype (Altınordu et al. Citation2016). The total genomic DNA was extracted for following the operation. The genomic DNA was manufactured to average 350 bp paired-end (PE) library using the Illumina Hiseq platform and sequenced using Illumina genome analyzer (Hiseq PE150). After comparing with all existing chloroplast genomes of Apioideae in GenBank, chloroplast genome-related reads of P. smithii were obtained. The raw reads were then assembled using NOVOPlasty 3.7 (Dierckxsens et al. Citation2017) with ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcL) gene from Angelica dahurica as seed file. The draft sequence obtained from NOVOPlasty was corrected subsequently using bowtie2 (Langmead and Salzberg Citation2012) and Tablet (Milne et al. Citation2013). After that, the complete genome sequence was annotated using Geneious 11.04 (Kearse et al. Citation2012) by pairwise align with the sequence of A. dahurica (GenBank accession number is KT963037).
Like other Apiaceae members, it also has two large inverted repeats (IRs), separated by large single-copy (LSC) and small single-copy (SSC) regions. The complete chloroplast genome of P. smithii (GenBank accession no. MN970011) was 147,130 bp in length, consisting of two IRs (18,264 bp) separated by the LSC (93,158 bp) and SSC (17,444 bp) regions. The genome contained 129 genes, including 85 protein-coding genes, 36 tRNA genes, and 8 rRNA genes respectively. The GC-content of the complete chloroplast genome was 37.5%.
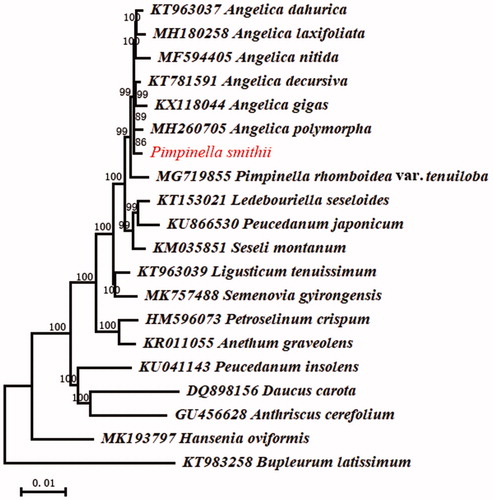
To ascertain the phylogenetic relationship between P. smithii and other species of Apiaceae, 19 cp genome sequences were achieved from the NCBI. All sequences were aligned using MAFFT v7 (Katoh and Standley Citation2013) and trimmed properly by trimAl (Capella-Gutierrez et al. Citation2009). Phylogenetic analysis was conducted using Maximum-Likelihood method using RAxML v8 (Stamatakis Citation2014) by applying the parameters ‘-f a -m GTRGAMMA -p 12345 -x 12345 -#1000’ where the GTRGAMMA model and 1000 bootstrap replicates were applied. Resembling the species P. rhomboidei var. tenuiloba (Tan and Yu Citation2018), P. smithii was closely allied with Angelica species (), which was consistent with the previous phylogenetic study using ITS and two cp introns sequences (Wang et al. Citation2014).
Acknowledgments
We deeply appreciate the constructive comments of Yan Yu on drafts of the manuscript. We are also extremely thankful to the anonymous reviewers that work in this paper.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Altınordu F, Peruzzi L, Yu Y, He X. 2016. A tool for the analysis of chromosomes: KaryoType. Taxon. 65(3):586–592.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 14(2):193–202.
- Pu FT, Watson MF. 2005. Pimpinella L. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 14. Beijing: Science Press and St. Louis: Missouri Botanical Garden Press; p. 93–104.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tan JB, Yu Y. 2018. The complete chloroplast genome of Pimpinella rhomboidea var. Tenuiloba. Mitochondrial DNA B. 3(1):101–102.
- Wang ZX, Downie SR, Tan JB, Liao CY, Yu Y, He XJ. 2014. Molecular phylogenetics of Pimpinella and allied genera (Apiaceae), with emphasis on Chinese native species, inferred from nrDNA ITS and cpDNA intron sequence data. Nordic J Bot. 32(5):642–657.