Abstract
The complete mitochondrial genome of Choroterpides apiculata (Ephemeroptera: Leptophlebiidae) is typically a circular molecule of 15,199 bp in length, containing 37 genes (13 protein-coding genes, 22 tRNAs, and two rRNAs) and one control region. The overall A + T content of the whole genome is 74% and the A + T content of the control region is 79.7%. In Bayesian inference and maximum-likelihood phylogenetic trees using 24 species from 13 families of Ephemeroptera, the monophyly of the families Isonychiidae, Heptageniidae, Vietnamellidae, Leptophlebiidae, Caenidae, and Baetidae were highly supported and C. apiculata was a sister group to Habrophlebiodes zijinensis.
Species of the Leptophlebiidae family (Insecta: Ephemeroptera) are distributed all over the world, including in China. The phylogenetic relationships of Ephemeroptera are still disputed based on morphological and/or molecular studies (Hebert et al. Citation2003; Ogden and Whiting Citation2005; Zhang et al. Citation2008; Ogden et al. Citation2009; Simon and Hadrys Citation2013; Li et al. Citation2014; Misof et al. Citation2014; Cai et al. Citation2018). There was only one mitogenome of species (Habrophlebiodes zijinensis) in Leptophlebiidae and so its monophyly could not be tested. Thus, it is necessary to sequence more mitogenomes of Leptophlebiidae species to discuss its phylogenetic relationship within Ephemeroptera.
Samples of Choroterpides apiculata were collected from Jinggu (23°57’38” N, 100°51’47” E), Yunnan province, China on 28 August Citation2018 and identified by Dr. JY Zhang. The sample (YN20180828) was identified and stored at a −40 °C freezer in the Animal Specimen Museum, College of Life Sciences and Chemistry, Zhejiang Normal University, China. Total genomic DNA was extracted from leg tissue of individual samples using Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China) and stored in the Zhang’s laboratory. Universal primers (Wang et al., Citation2019; Zhang, Cai, et al. Citation2018; Zhang, Yu, et al. Citation2018) were used to amplify some partial fragments. The mitogenome was deposited in GenBank with the accession number MN807287.
The complete mitogenome of C. apiculata was a circular molecule of 15,199 bp in length, including 13 protein-coding genes (PCGs), 22 tRNAs, two rRNAs, and one control region. The gene arrangement and orientation coincided with those of typical mayfly species. The overall base composition of the major strand was as follows: A (34.5%), T (39.5%), G (10.1%), and C (15.9%). All 13 PGCs used the standard mitochondrial start codon ATN. Eleven PCGs terminated with a complete stop codon (TAA), whereas COX2 and ND4 showed an incomplete terminal T– –.
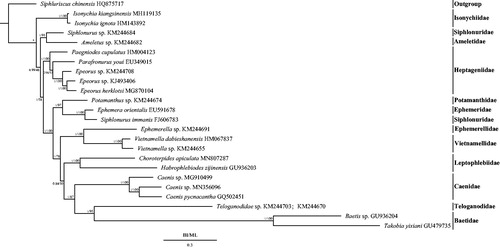
Phylogenetic trees were constructed from the 13 PCGs using two methods: Bayesian inference (BI) using MrBayes 3.1.2 (Huelsenbeck and Ronquist Citation2001) and maximum-likelihood (ML) using RAxML 8.2.0 (Stamatakis Citation2014). Siphluriscus chinensis was used as the outgroup according to the method of Li et al. (Citation2014) and Xu et al. (Citation2020). The mitogenomes of 23 Ephemeroptera species were downloaded from GenBank (Zhang et al. Citation2008; Li et al. Citation2014; Tang et al. Citation2014; Zhou et al. Citation2016; Cai et al. Citation2018; Gao et al. Citation2018; Ye et al. Citation2018; Xu et al. Citation2020) and used to investigate the phylogenetic relationships. In order to select the conserved regions of nucleotides, each alignment was performed by Gblock 0.91 b (Castresana Citation2000) using the default settings. In BI and ML phylogenetic trees, the monophyly of the families Isonychiidae, Heptageniidae, Vietnamellidae, Leptophlebiidae, Caenidae, and Baetidae were supported, whereas the monophyly of Siphlonuroidea failed (). Leptophlebiidae was supported as a sister group to the clade Caenidae + (Teloganodidae + Baetidae), which also agreed with the results of Cai et al. (Citation2018), Gao et al. (Citation2018), Ye et al. (Citation2018), and Xu et al. (Citation2020). Choroterpides apiculata was supported as the sister group to H. zijinensis.
Figure 1. Phylogenetic tree of the relationships among 24 species of Ephemeroptera, including Choroterpides apiculata (MN807287) based on the nucleotide dataset of the 13 mitochondrial protein-coding genes (PCGs). Siphluriscus chinensis was used as the outgroup. The numbers above branches specify posterior probabilities as determined from BI (left) and bootstrap percentages from ML (right). The GenBank numbers of all species are shown in the figure.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cai YY, Gao YJ, Zhang LP, Yu DN, Storey KB, Zhang JY. 2018. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) and the phylogeny of Ephemeroptera in Pterygota. Mitochondrial DNA B. 3(2):577–579.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Gao XY, Zhang SS, Zhang LP, Yu DN, Zhang JY, Cheng HY. 2018. The complete mitochondrial genome of Epeorus herklotsi (Ephemeroptera: Heptageniidae) and its phylogeny. Mitochondrial DNA B. 3(1):303–304.
- Hebert PDN, Cywinska A, Ball SL, Dewaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc Lond B. 270(1512):313–321.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Li D, Qin JC, Zhou CF. 2014. The phylogeny of Ephemeroptera in Pterygota revealed by the mitochondrial genome of Siphluriscus chinensis (Hexapoda: Insecta). Gene. 545(1):132–140.
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science. 346(6210):763–767.
- Ogden TH, Gattolliat JL, Sartori M, Taniczek AH, Soldán T, Whiting MF. 2009. Towards a new paradigm in mayfly phylogeny (Ephemeroptera): combined analysis of morphological and molecular data. Syst Entomol. 34(4):616–634.
- Ogden TH, Whiting MF. 2005. Phylogeny of Ephemeroptera (mayflies) based on molecular evidence. Mol Phylogenet Evol. 37(3):625–643.
- Simon S, Hadrys H. 2013. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol Phylogenet Evol. 69(2):393–403.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tang M, Tan MH, Meng GL, Yang SZ, Su X, Liu SL, Song WH, Li YY, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes – a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42(22):e166.
- Wang J, Dai XY, Xu XD, Zhang ZY, Yu DN, Storey KB, Zhang JY. 2019. The complete mitochondrial genomes of five longicorn beetles (Coleoptera: Cerambycidae) and phylogenetic relationships within Cerambycidae. PeerJ. 7:e7633.
- Xu XD, Jia YY, Dai XY, Ma JL, Storey KB, Zhang JY, Yu DN. 2020. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) from Fujian and the phylogeny of Caenidae within Ephemeroptera. Mitochondrial DNA B. 5:152–153.
- Ye QM, Zhang SS, Cai YY, Storey KB, Yu DN, Zhang JY. 2018. The complete mitochondrial genome of Isonychia kiangsinensis (Ephemeroptera: Isonychiidae). Mitochondrial DNA B. 3(2):541–542.
- Zhang JY, Zhou CF, Gai YH, Song DX, Zhou KY. 2008. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene. 424(1-2):18–24.
- Zhang LP, Cai YY, Yu DN, Storey KB, Zhang JY. 2018. Gene characteristics of the complete mitochondrial genomes of Paratoxodera polyacantha and Toxodera hauseri (Mantodea: Toxoderidae). PeerJ. 6:e4595.
- Zhang LP, Yu DN, Storey KB, Cheng HY, Zhang JY. 2018. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera, Mantodea) and the phylogeny within Mantodea. Int J Biol Macromol. 111:787–795.
- Zhou D, Wang YY, Sun JZ, Han YK, Zhou CF. 2016. The complete mitochondrial genome of Paegniodes cupulatus (Ephemeroptera: Heptageniidae). Mitochondrial DNA A. 27(2):925–926.
