Abstract
The Apteryx rowi has been listed in the International Union for Conservation of Nature (IUCN) Red List of Threatened Species due to a population combination of habitat loss and predation by introduced mammals. Mitochondrial genome sequence is informative and very useful for systematics and conservation biology studies. Here, we first reported the complete mitochondrial genome sequence of A. rowi by next-generation sequencing (NGS). The complete mitochondrial genome sequence of A. rowi was 17,303 bp in length and contained 13 protein-coding genes (PGCs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA genes, and 1 control region (D-loop). The overall nucleotide composition of this mitochondrial genome was 31.07% A, 29.43% C, 26.38% T, and 13.12% G, respectively, and the percentage of G + C content was 42.55%. This result provided new molecular biology information to further understand the genetic diversity of the A. rowi, which will help to protect this population.
The Apteryx rowi belongs to the genus Apteryx within the family Apterygidae. The Apteryx rowi (Okarito brown kiwi), known as its shrill ‘keee WEEE’. This species is distributed widely in New Zealand, Okarito, Westland (Robertson and de Monchy Citation2012), they have been recently introduced to Mana, Motuara and Blumine Islands, in the Cook Strait region, and breeding has been recorded at both Mana and Blumine (Robertson et al. Citation2017). Apteryx rowi has been listed in the IUCN Red List of Threatened Species and included in the ‘Washington Convention’ CITES I level protected animals (BirdLife International, Citation2014). However, the total population remains small and it is therefore classified as Vulnerable Molecular. Molecular studies about A. rowi are limited and the complete mitochondrial genome of A. rowi has not been determined and characterized until now. Here, we assembled and characterized the complete mitochondrial genome of A. rowi.
The total genomic DNA was extracted from the blood of adult male New Zealand (S34°47′, E166°178′) and the whole-genome library of genomic DNA was sequenced using the Illumina Hiseq2500 platform. The information of sample of A. rowi was stored in NCBI (Accession No. SRR6918118). The sequencing reads were trimmed using Trimmomatic v0.36 (Bolger et al. Citation2014), and assembled with mira v4.0.2 and MITObim v1.8 software (Hahn et al. Citation2013), then the 37 genes present in the genome were annotated using DOGMA (Wyman SK et al. Citation2004) and tRNAscan-SE 2.0 (http://lowelab.ucsc.edu/tRNAscan-SE/).
The complete mitochondrial genome of A. rowi is a double-stranded, circular DNA 17,303 bp in total length (GenBank Accession No. MN998652). The complete mitochondrial genome of A. rowi has a length of 17,303 bp with a typical circle structure that contains 22 tRNA genes, 13 protein-coding genes, and 2 rRNA genes.
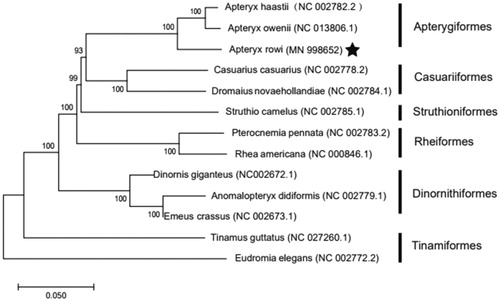
The phylogenetic analysis of 13 mitochondrial genomes using MEGA 7(Kumar et al. Citation2016) showed that A. rowi and Apteryx owenii are the most closely related species (). The mitogenome of A. rowi would contribute to the understanding of the phylogeny and evolution of Apterygidae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- BirdLife International. 2014. Apteryx rowi. The IUCN Red List of Threatened Species. Version 2014.2. [accessed 2014 Oct 21]. http://www.jucnredlist.org.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 30(15):2114–2120.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Kumar S, Stecher G, Tamura K. 2016. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Robertson HA, Baird K, Dowding JE, Elliott GP, Hitchmough RA, Miskelly CM, McArthur N, O’Donnell CFJ, Sagar PM, Scofield RP, et al. 2017. Conservation status of New Zealand birds, 2016. New Zealand Threat Classification Series 19. Wellington, Department of Conservation. 27p.
- Robertson HA, de Monchy PJM. 2012. Varied success from the landscape-scale management of kiwi Apteryx spp. in five sanctuaries in New Zealand. Bird Conserv Int. 22(4):429–444.
- Wyman SK, Jansen RK, Boore JL. 2004. DOGMA citation: automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.