Abstract
The complete mitogenome of Pseudotolithus typus (Perciformes: Sciaenidae) is analyzed by high-throughput sequencing. The 16,502 nucleotides of assembled mitogenome has 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and the 1 putative control region. Incomplete stop codons were found in seven genes (ND2, ND3, ND4, COII, COIII, CytB, ATPase6). The phylogenetic analysis indicated that P. typus is closely related to P. elongatus. This study will provide useful information to understand the evolutionary and phylogenetic exploration of the family Sciaenidae.
Longneck croaker Pseudotolithus typus, one of the important imported fish in Korea, is commonly inhabited in the eastern Atlantic (Chao and Trewavas Citation1990). Since longneck croaker has a very similar taste and shape to small yellow croaker (Larimichthys polyactis) that Koreans like to eat, its import to Korea is gradually increasing. Recently, cases of misconduct sales using these similarities have been rapidly increasing, and it is serious to develop a discrimination method to prevent these. Not only biological studies but also genetic researches on longneck croaker are not abundant to use for this purpose. For this reason, objective of this study is to make the genetic data available for longneck croaker through sequencing and characterizing the mitogenome.
The specimen was obtained from a global fish importer in Korea. It was caught in the Guinea coast (9°40′54.5″N 14°50′10.7″ W) and carried to Korea in the frozen state. The specimen is now deposited at the Department of Food Engineering, Pukyong National University (Sample no. MFDS-FMI07) under the research funding from the Ministry of Food and Drug Safety. The extracted total genomic DNA was used for the construction of DNA library using the TruSeq Nano DNA Sample Preparation kit (Illumina, San Diego, CA). The constructed library was sequenced by Illumina NextSeq 500 with paired-ends. The cleaned sequences were mapped to previously reported mitogenome of P. elongatus (Kim et al. Citation2019) using Geneious 11.1.3 (Kearse et al. Citation2012). The annotation was accomplished by MitoFish pipeline (Sato et al. Citation2018), and the phylogenetic tree was illustrated using MEGA X (Kumar et al. Citation2018) with Maximum Likelihood methods.
The 16,502 nucleotides of assembled mitogenome (GenBank accession no. MN973879) has 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 putative control region. The overall base composition of the mitogenome was estimated as follows: A 27.1%, T 25%, G 16.5% and C 31.4%, with an A + T contents 52.1%. Incomplete stop codons were found in seven genes (ND2, ND3, ND4, COII, COIII, CytB, ATPase6) and atypical codon usages were found in tRNA-Leu (UAA, UAG), tRNA-Ser (UGA, GCU). Only ND6 gene was located on the L-strand among the protein-encoding genes while the eight tRNA (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro) genes were positioned on the H-strand.
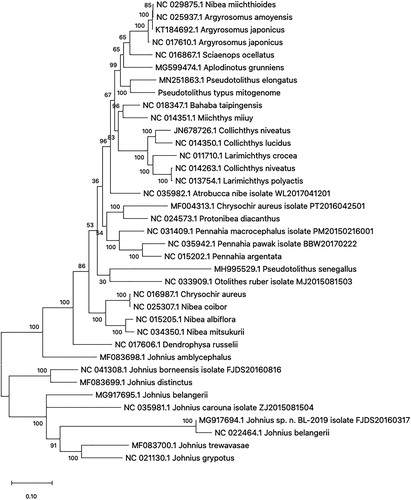
The phylogenetic analysis of 37 mitogenomes in Sciaenidae indicated that P. typus is strictly related to the Pseudotolithus elongatus (Accession No. MN251863) while P. senegallus (Accession No. MH995529) is having far genetic distance even the species that are in the same genus (). This study will provide useful information to understand the evolutionary and phylogenetic exploration of the family Sciaenidae and the genus Pseudotolithus.
Figure 1. Phylogenetic tree of 37 species in family Sciaenidae. The 36 mitogenome sequences from GenBank database and Pseudotolithus typus mitogenome sequence were aligned using ClustalW and phylogenetic analysis was conducted by maximum likelihood method with 1000 bootstrap. The percentage at each node is the bootstrap probability.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chao LN, Trewavas E. 1990. Sciaenidae. In: Quero JC, Hureau C, Karrer AP, Saldanha L, editors. Check-list of the fishes the eastern tropical Atlantic (CLOFETA). Lisbon: JNICT; Paris: SEI, UNESCO; Vol. 2, p. 813–826.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim JO, Seo YB, Shin JY, Yang JY, Kim GD. 2019. The complete mitochondrial genome of bobo croaker Pseudotolithus elongatus (Perciformes: Sciaenidae). Mitochondrial DNA Part B. 4(2):3179–3181.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Bio Evol. 35(6):1547–1549.
- Sato Y, Miya M, Fukunaga T, Sado T, Iwasaki W. 2018. MitoFish and MiFish Pipeline: a mitochondrial genome database of fish with an analysis pipeline for environmental DNA metabarcoding. Mol Bio Evol. 35(6):1553–1555.
