Abstract
We have sequenced the female type (F-type) complete mitochondrial genomes of two Hyriposis species, H. schlegelii and H. cumingii (Gonideinae, Unionidae, Unionida, Bivalvia) from Lake Kasumigaura, Japan, and inferred the Unioninae phylogeny. Complete mitochondrial genomes (H. schlegelii, 15,954 bp, LC498622; H. cumingii, 15,961 bp, LC498621) contain 13 protein-coding genes (PCGs), 2 rRNA genes, and 22 tRNA genes. Molecular phylogenetic analyses using the 13 PCGs including the two species were performed. This study should be basic data to investigate the evolution of Gonideinae and genetic diversity of Hyriposis species in local populations.
Hyriopsis schlegelii is an endangered species, endemic to Lake Biwa, Japan. Since the 1930s, it was transplanted to Lake Kasumigaura, Japan. Hyriopsis cumingii inhabits in China, and produces freshwater pearls (Wei et al. Citation2016). This species was brought to Lake Kasumigaura from the 1970s to 1980s. Since the two species inhabited sympatrically, hybridization between the two has been suspected (Shirai et al. Citation2010).
To obtain molecular basis for the conservation of the two species in Lake Kasumigaura, we have sequenced complete mitochondrial genomes of H. schlegelii and H. cumingii collected at Lake Kasumigaura and molecular phylogenetic analyses of Gonideinae were performed.
Hyriopsis schlegelii and H. cumingii (#UU-SBD-Unio-10 and 11, respectively) were collected at Lake Kasumigaura, Japan (N35.992, E140.349). DNA extraction and sequence analyses were performed as described by Fukata and Iigo (Citation2019). Local BLAST search using the mitochondrial genome of H. schlegelii (AP018551) and H. cumingii (HM347668) identified the F-type complete mitochondrial genomes of the two species (circular; H. schlegelii: 15,954 bp: H. cumingii, 15,961 bp). The complete mitogenomes contain 13 protein-coding genes (13PCGs), 2 rRNA genes, and 22 tRNAs. The sequences have been submitted to DDBJ/EMBL/Genbank with accession numbers LC498622 (H. schlegelii) and LC498621 (H. cumingii).
Male and female mitogenomes of Gonideinae are known to have different gene orders (Breton et al. Citation2009). The addition of M-type sequences to the traditional F-type mitogenomes could often result in more robust evolutionary relationships than those provided by the F-type only (Walker et al. Citation2006). As expected, molecular phylogenetic tree constructed by MEGA X (Kumar et al. Citation2018) using the 13 PCGs of Gonideinae revealed two main clades (F-type and M-type; ). Based on the tree, the three mitogenomes (NC_015110, HQ641407 and NC_011763) seem to have been misidentified: NC_011763 (H. cumingii) might be H. schlegelii; NC_015110 and HQ641407 (both registered as H. schlegelii) might be the F-type mitogenomes of a new Hyriposis species (identity of nucleotides and deduced amino acid sequences to the F-type H. cumingii and H. schlegelii range from 94.0 to 94.8 and 89.3 to 90.6%, respectively).
Figure 1. Molecular phylogenetic tree (Maximum likelihood method) using 13 protein-coding genes of Gonideinae including H. schlegelii and H. cumingii from Lake Kasumigaura, Japan. The numbers above the branch meant bootstrap value (1000 replicates). Leaf names were presented as species names and accession number. Sequences determined in this study (LC498621 and LC498622) are in bold. Sequences thought to be misidentified are underlined.
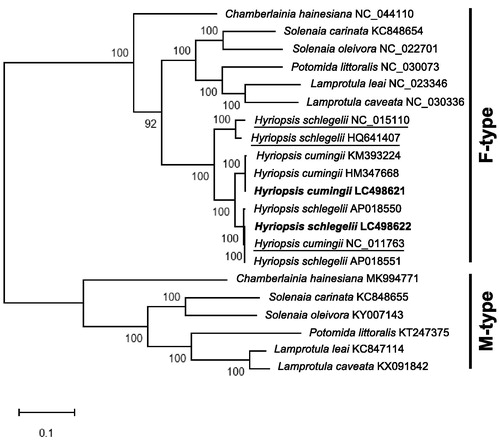
This research would provide valuable information for investigating the evolution of Gonideinae and the genetic diversity of local populations of Hyriposis species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Breton S, Beaupre HD, Stewart DT, Piontkivska H, Karmakar M, Bogan AE, Blier PU, Hoeh WR. 2009. Comparative mitochondrial genomics of freshwater mussels (Bivalvia: Unionoida) with doubly uniparental inheritance of mtDNA: gender-specific open reading frames and putative origins of replication. Genetics. 183(4):1575–1589.
- Fukata Y, Iigo M. 2019. The complete mitochondrial genome of freshwater mussel Nodularia douglasiae (Unionidae) from Lake Kasumigaura, Japan, and its phylogenetic analysis. Mitochondrial DNA B. 4(2):3488–3489.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Shirai A, Kondo T, Kajita T. 2010. Molecular markers reveal genetic contamination of endangered freshwater pearl mussels in pearl culture farms in Japan. Venus. 68:151–163.
- Walker JM, Bogan AE, Bonfiglio EA, Campbell DC, Christian AD, Curole JP, Harris JL, Wojtecki RJ, Hoeh WR. 2006. Primers for amplifying the hypervariable, male-transmitted COII-COI junction region in amblemine freshwater mussels (Bivalvia: Unionoidea: Ambleminae). Mol Ecol Notes. 7(3):489–491.
- Wei M, Yang S, Yu P, Wan Q. 2016. The complete mitochondrial genome of Hyriopsis cumingii (Unionoida: Unionidae): genome description and related phylogenetic analyses. Mitochondrial DNA A. 27:1769–1770.
