Abstract
The complete mitochondrial genome (mitogenome) of a Satirid species Coenonympha amaryllis was assembled and annotated. The mitogenome is a DNA molecule of 15125 bp, and consists of 13 protein-coding genes (PCGs), 22 transfer RNAs, 2 rRNAs, and 1 A–T rich region. The nucleotide composition biases toward A and T is 79.4% of the entirety, which is a typical structure of Lepidopterans. All PCGs started with ATN, except cox1, which started with CGA, 8 PCGs stop with TAN and 5 genes exhibited incomplete stop codon. Phylogenetic analysis revealed that Satyrinae is a monophyletic group and Coenonympha (C. amaryllis) as sister of the genus Triphysa (T. phryne).
Keywords:
Satyrinae, one of the 12 subfamilies of Nymphalidae, is highly diverse and distributed worldwide, including ∼2500 described species (Peña and Wahlberg Citation2008). However, the phylogenetic relationships of Satyrinae have no robust hypotheses (Yang and Zhang Citation2015). Recent works show mitogenome is effective as molecular markers in phylogeny analysis in Lepidopterans (Kim et al. Citation2014; Timmermans et al. Citation2016; Yang M. et al. Citation2019; Yang Z. et al. Citation2019)
In this study, we collected samples of Coenonympha amaryllis from Shaanxi province, China (E109.37, N36.03) in August 2019. Specimen (Voucher No. LE250372) was deposited in the Entomological Museum of Northwest A&F University, Shaanxi province, China. Total genome DNA was extracted from the thorax muscle of a male adult. The complete mitogenome of C. amaryllis is a circular molecule of 15125 bp in size (Genbank accession number is MN756798), including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 A–T rich region (Control region). Its gene arrangement and direction are in accordance with other Nymphalid butterflies (Xu et al. Citation2019). The C. amaryllis mitogenome has an A–T content of 79.4% (A 38.1%; T 41.3%; C 12.6%; G 8.0%), which is a typical structure of Lepidopterans mitogenome. Generally, PCGs are started with ATN codon and terminated with TAN, but one gene (cox1) is initiated by CGA, five PCGs (cox1, cox2, nd1, nd4 and nd5) are stopped with single-nucleotide (T––), which was reported in another study (Shi et al. Citation2019). All tRNA genes are identified by MITOS web server (Bernt et al. Citation2013). The 16S rRNA gene is 1337 bp in size and is located between tRNA-L1 and tRNA-V; the 12S rRNA gene is 759 bp in length and is located between tRNA-V and A–T-rich region. The control region is 308 bp long, and is located between 12S rRNA and tRNA-M.
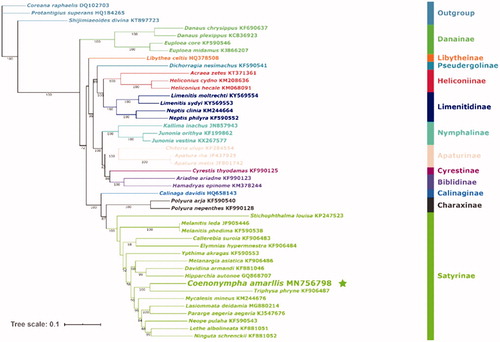
Here, we reconstructed a phylogeny of Nymphalidae using 13 PCGs from C. amaryllis and other 41 representatives () from 12 subfamilies of the family and 3 outgroup species () from Lycaenidae. Thirteen concatenated PCG sequences of mitogenomes were analyzed by maximum likelihood (ML) in IQ-tree (Lam-Tung et al. Citation2015). The phylogenetic tree showed that Satyrinae was a monophyletic group in Nymphalidae and Coenonympha (C. amaryllis) was sister of the genus Triphysa (T. phryne).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogen Evol. 69(2):313–319.
- Kim MJ, Wang AR, Park JS, Kim I. 2014. Complete mitochondrial genomes of five skippers (Lepidoptera: Hesperiidae) and phylogenetic reconstruction of Lepidoptera. Gene. 549(1):97–112.
- Lam-Tung N, Schmidt HA, von Haeseler A, Bui Quang M. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Peña C, Wahlberg N. 2008. Prehistorical climate change increased diversification of a group of butterflies. Biol Lett. 4(3):274–278.
- Shi QH, Lin XQ, Ye X, Xing JH, Dong GW. 2019. Characterization of the complete mitochondrial genome of Minois dryas (Lepidoptera: Nymphalidae: Satyrinae) with phylogenetic analysis. Mitochondrial DNA Part B. 4(1):1447–1449.
- Timmermans M, Lees DC, Thompson MJ, Safian S, Brattstrom O. 2016. Mitogenomics of ‘Old World Acraea’ butterflies reveals a highly divergent ‘Bematistes’. Mol Phylogen Evol. 97:233–241.
- Xu C, Pan ZQ, Nie L, Hao JS. 2019. The complete mitochondrial genome of Issoria eugenia (Lepidoptera: Nymphalidae: Heliconiinae). Mitochondrial DNA Part B. 4(1):1662–1663.
- Yang M, Song L, Shi YX, Li JH, Zhang Y, Song N. 2019. The first mitochondrial genome of the family Epicopeiidae and higher-level phylogeny of Macroheterocera (Lepidoptera: Ditrysia). Int J Biol Macromol. 136:123–132.
- Yang Z, Yang TT, Liu Y, Zhang HB, Tang BP, Liu QN, Ma YF. 2019. The complete mitochondrial genome of Sinna extrema (Lepidoptera: Nolidae) and its implications for the phylogenetic relationships of Noctuoidea species. Int J Biol Macromol. 137:317–326.
- Yang M, Zhang Y. 2015. Phylogenetic utility of ribosomal genes for reconstructing the phylogeny of five Chinese satyrine tribes (Lepidoptera, Nymphalidae). Zookeys. 488:105–120.