Abstract
In this research, we determined and described the complete mitochondrial genome of Black-streaked Scimitar Babbler (Pomatorhinus dedekeni) which was the first complete mitogenome reported. The complete genome of P. dedekeni was 16,996 bp in length and contained 13 protein-coding genes, 22 transfer RNA genes, two ribosome RNA genes, and one non-coding control region. The overall base composition of the mitochondrial DNA was 29.2% for A, 22.9% for T, 14.6% for G, and 33.3% for C, with a GC content of 47.9%. The information of P. dedekeni will contribute to the phylogenetic studies of the Timaliidae.
Black-streaked Scimitar Babbler (Pomatorhinus dedekeni) is found in Burma, Laos, mainland China, and Vietnam (Birdlife International Citation2018). In China, P. dedekeni is distributed from eastern Tibet to western Sichuan (Birdnet Citation2014) and is called Rusty-cheeked Scimitar-Ba (Zheng Citation2006). The species was rated safe for conservation and was classified as the IUCN red list of endangered species in the least popular species (LC) (IUCN Citation2018). The habitat of P. dedekeni includes subtropical or tropical (lowland) moist shrub, subtropical or tropical (lowland) dry shrub, subtropical or tropical moist lowland forest, subtropical or tropical high altitude sparse shrub, subtropical or tropical (lowland) steppe, and subtropical or tropical moist mountain forest (Birdnet Citation2014). There are eight subspecies of Black-streaked Scimitar Babbler, decarlei, dedekeni, odicus, abbreviatus, swinhoei, erythrocnemis, cowensae, and gravivox (Birdnet Citation2014). However, there is little molecular data reported on the subspecies of Pomatorhinus gravivox. Therefore, in order to better understand the phylogeny and evolution of this species or subspecies in the Timaliidae family, we sequenced the complete mitochondrial genome of P. dedekeni.
A natural dead P dedekeni was collected from Xiejia village, Marong township, Baiyu county, Ganzi Tibetan Autonomous region, Sichuan province. The specimen is now in the Key Laboratory of Bioresources and Ecoenvironment, Sichuan University. The stored number of the sample is 2019PD-1. Total genomic DNA was extracted from muscle tissue. The complete mitochondrial genome was amplified using PCR and PCR produces were sequenced using ABI PRISM 3730 DNA sequencer. The complete mitochondrial genome of P. dedekeni was submitted to the NCBI database under the accession number MN918717. The full genome of P. dedekeni mitochondrial is a circular DNA molecule and is 16,996 bp in length. The overall base composition was 29.2% A, 22.9% T, 14.6% C, and 33.3% G, with a GC ratio of 47.9%. The complete mitogenome of P. dedekeni included 13 protein-coding genes, two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA (tRNA) genes, and one non-coding control region (D-loop). Among these genes, ND6 and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, and tRNA-Pro) located on the light strand (L-strand), whereas other genes located on the heavy strand (H-strand). The insertion of an extra cytosine at position 147 of the ND3 protein-coding sequence and pseudo-control region as in many birds (Song et al. Citation2015; Yan et al. Citation2017; Jiang et al. Citation2019) were not found in P. dedekeni mitogenome.
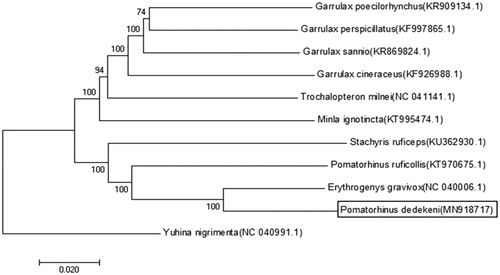
The reconstructed phylogenetic tree supports the position of P. dedekeni in the Timaliidae (). All nodes are strongly supported by maximum reduction and Bayesian posterior probability analysis. As indicated by the trees (), the result is similar to the previous study on another subspecies, P. gravivox (Duan et al. Citation2019). The sequence data will be useful for the phylogenetic studies of the family Timaliidae species in the future.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Birdlife International. 2018. Species factsheet: Pomatorhinus gravivox. [downloaded 2018 Nov 30]. http://www.birdlife.org.
- Birdnet. 2014. Spot-breated Scimitar Babbler. www.cnniao.com/niao/1604.html.
- Duan YB, Li Y, Luo X. 2019. Next-generation sequencing yields the mitochondrial genome of black-streaked Scimitar Babbler Pomatorhinus gravivox (Passeriformes:Timaliidae). Mitochondrial DNA B. 4(1):998–999.
- IUCN. 2018. The IUCN Red List of Threatened Species. Version 2018-1. [accessed 2018 Nov 20]. www.incnredlist.org.
- Jiang X, Gao J, Zhou C, Jing J, Chen ML, Yue B, Zhang XY. 2019. The complete mitochondrial genome of brown-flanked bush warbler (Horornis fortipes). Mitochondrial DNA B. 4(1):1426–1427.
- Song XH, Huang J, Yan CC, Xu GW, Zhang XY, Yue BS. 2015. The complete mitochondrial genome of Accipiter virgatus and evolutionary history of the pseudo-control regions in Falconiformes. Biochem Syst Ecol. 58:75–84.
- Yan C, Mou B, Meng Y, Tu F, Fan Z, Price M, Yue BS, Zhang XY. 2017. A novel mitochondrial genome of Arborophila and new insight into Arborophila evolutionary history. PLoS One. 12(7):e0181649.
- Zheng ZX. 2006. Latin, Chinese and English comparison of world bird names. Beijing: Science Press.