Abstract
Phaeodactylum tricornutum ICE-H is a single-cell eukaryotic alga that can grow in the extreme Antarctic environment. The complete chloroplast genome of Phaeodactylum tricornutum ICE-H was assembled with the Illumina sequencing. The chloroplast genome was 117,363 bp in size, containing a large single-copy region (LSC, 63,668 bp), a small single-copy region (SSC, 39,871 bp), and a pair of inverted repeat regions (IRs, 13,824 bp each). The overall GC content was 32.15%. A total of 168 genes were predicted including 132 protein-coding genes, 30 tRNAs, and 6 rRNAs. Phylogenetic analysis indicated that Phaeodactylum tricornutum ICE-H was closely related to Didymosphenia geminata.
Antarctic sea ice microalgae have a crucial effect on maintaining the stability of polar ecosystems (Thomas and Dieckmann Citation2002). They are important primary producers of the Southern ocean, which provide important food sources for zooplankton, krill, and fish (Lizotte Citation2001; Qu et al. Citation2017). Meanwhile, Antarctic sea ice microalgae contain many active substances which have great potential to be exploited as biotechnological products (Zuliani et al. Citation2016). Phaeodactylum tricornutum ICE-H, as an important eukaryotic alga, belongs to Phaeodactylaceae and can prosperously grow in the extreme Antarctic environment. Therefore, there is an immediate need to understand the genomic information of the Phaeodactylum tricornutum ICE-H. Here, we firstly assembled and annotated the complete chloroplast genome of Phaeodactylum tricornutum ICE-H.
Total chloroplast DNA was harvested with the SDS method from the P. tricornutum ICE-H collected from ice floes near the Zhongshan Research Station, Antarctica (69°S, 77°E) (Lim et al. Citation2016). The specimen (Accession no. FIO2008697701) was stored in the Key Laboratory of Marine Bioactive Substances, the First Institute of Oceanography, Ministry of Natural Resource, China. The whole genome was sequenced with the Illumina NovaSeq PE150 at the Beijing Novogene Bioinformatics Technology Co., Ltd. Additionally, SPAdes software was used to assemble the complete circular chloroplast genome. The complete genome was annotated with GeneMarkS software (Besemer et al. Citation2001).
The total size of the circular chloroplast genome was 117,363 bp with 32.15% GC content (Genbank accession number: MN937452). The large single-copy region (LSC, 63,668 bp) and the small single-copy region (SSC, 39,871 bp) were separated by a pair of reverse repeat regions (IRs, 13,824 bp) regions. There are 168 genes in the chloroplast genome contained 132 protein-coding genes, 30 tRNA genes, and 6 rRNA genes.
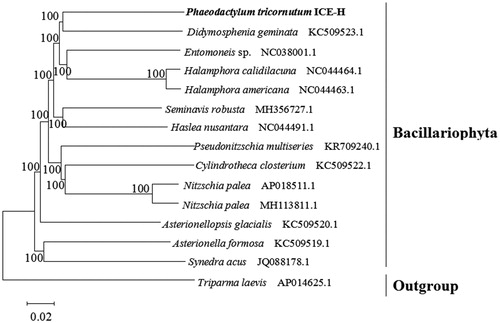
To ascertain the phylogenetic position of P. tricornutum ICE-H, a phylogenetic analysis was conducted with 15 complete chloroplast genomes. Fourteen species belonged to Bacillariophyta and Triparma laevis was used as an outgroup. The phylogenetic tree was constructed with MEGA 7.0 and the setting of bootstraps was 1000 (Kumar et al. Citation2016). The result revealed that P. tricornutum ICE-H was closely related to Didymosphenia geminata (). Our findings will provide a basis for further study on the evolution of the Bacillariophyta chloroplast genome.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29(12):2607–2618.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lim HJ, Lee EH, Yoon Y, Chua B, Son A. 2016. Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J Appl Microbiol. 120(2):379–387.
- Lizotte MP. 2001. The contributions of sea ice algae to Antarctic marine primary production. Am Zool. 41:57–73.
- Qu CF, Liu FM, Zheng Z, Wang YB, Li XG, Yuan HM, Li N, An ML, Wang XX, He YY, et al. 2017. Effects of ocean acidification on the physiological performance and carbon production of the Antarctic sea ice diatom Nitzschia sp. ICE-H. Mar Pollut Bull. 120(1–2):184–191.
- Thomas DN, Dieckmann GS. 2002. Antarctic Sea ice–a habitat for extremophiles. Science. 295(5555):641–644.
- Zuliani L, Frison N, Jelic A, Fatone F, Bolzonella D, Ballottari M. 2016. Microalgae cultivation on anaerobic digestate of municipal wastewater, sewage sludge and agro-waste. Int J Mol Sci. 17(10):1692.