Abstract
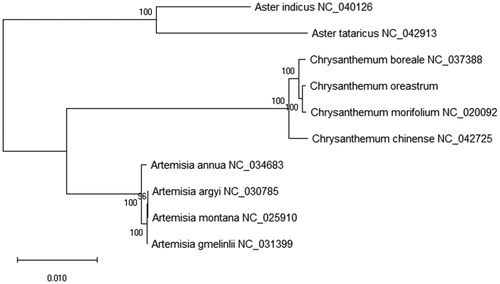
Chrysanthemum oreastrum is a perennial herb of Compositae and one of the traditional Chinese medicinal herb in China. In this study, the complete chloroplast genome of C. oreastrum was illustrated to add more genetic information. The chloroplast genome is 151,045 bp in length as the circular, which consists a large single-copy region (LSC) of 82,793 bp, a small single-copy region (SSC) of 18,348 bp and a pair of inverted-repeat regions (IRs) of 24,952 bp. The overall nucleotide composition of chloroplast genome is: 47,014 bp A (31.1%), 47,399 bp T (31.4%), 27,963 bp C (18.5%), 28,699 bp G (19.0%), and the total G + C content of 37.5%. 128 species genes were annotated that included 85 protein-coding genes (PCGs), 35 transfer RNA (tRNAs), and 8 ribosome RNA (rRNAs). Phylogenetic relationship indicated that C. oreastrum is closely related to C. morifolium in the genetic relationship constructed using the maximum-likelihood (ML) method.
Chrysanthemum oreastrum, native to Asia and northeastern Europe,is one of the most important and popular ornamentals over the world. It is also important as health foods and as anti-inflammatory herbs in Traditional Chinese Medicine. Recent pharmacological studies have focused on their various biological effects including antioxidative, cardiovascular protective, vasorelaxant, and anti-human immune deficiency virus activities (Song et al. Citation2018). Most chrysanthemum cultivars are polyploidy (2n = 4× = 36 or 2n = 6× = 54) and highly heterozygous (Xu et al. Citation2013). Now, the research of Traditional Chinese Medicine (TCM) has become a hot study direction. But, few genome information of TCM and publication that limited further research on TCM have been analysed. The chloroplast genome information of C. oreastrum can provide more data to study the evolution of this species. For this reason, we have studied the chloroplast genome of C. oreastrum that can be useful for phylogenetic relationship and implications research, which can also be used to provide basic data for the development of TCM in the future.
Using the plant tissues genomic DNA Extraction Kit (TIANGEN, BJ and CN) method, total genomic DNA was isolated from the fresh flowers of C. oreastrum and collected from herb market near Zhejiang Chinese Medical University that was located at Hangzhou, Zhejiang, China (30.09 N, 119.89E). The chloroplast genome DNA was stored in Zhejiang Chinese Medical University (No. SCMC-ZJU-TCM-08). It was purified and sequenced by the sequencer that collected raw sequences which were quality controlled and removed by the FastQC (Andrews Citation2015). The chloroplast genome of C. oreastrum was assembled and annotated by the MitoZ (Meng et al. Citation2019). The chloroplast genome map was generated by the OrganellarGenomeDRAW (Lohse et al. Citation2013). The annotated chloroplast genome sequence was submitted to the NCBI GenBank accession KX5657831.
The information of the chloroplast genome of C. oreastrum is obtained from NCBI GenBank No. JQ362483 as a reference sequence, which is 151,045 base pairs (bp) long and has a typical quadripartite structure, which consists of a large single-copy region (LSC of 82,781 bp), a small single-copy region (SSC of 18,348 bp), and a pair of inverted repeat regions (every IRs of 24,952 bp). The overall nucleotide composition of chloroplast genome is: 47,014 bp A (31.1%), 47,399 bp T (31.4%), 27,963 bp C (18.5%), 28,669 bp G (19.0%), and the total G + C content of 37.5%. The chloroplast genome of C. oreastrum contains 128 genes, which included 85 protein-coding genes (PCG), 35 transfer RNA genes (tRNAs), and 8 ribosomal RNA genes (rRNAs). Eighteen genes were found duplicated in IR regions, which included 7 PCGs species (rpl2, rpl23, ycf2, ndhB, rps7, rps12 and ycf1), 7 tRNAs species (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG and trnN-GUU), and 4 rRNAs species (rRNA16, rRNA23, rRNA4.5 and rRNA5).
To study the phylogenetic relationship of C. oreastrum with other nine plants species, chloroplast genomes were used to construct the phylogenetic tree using the maximum-likelihood (ML) method. The phylogenetic tree of ML analysis was performed using the MEGA X (Kumar et al. Citation2018) with the best model and all of the nodes were inferred with strong support by 2000 bootstrap values replicate for each node. The phylogenetic tree was drawn using the MEGA X and edited using the Evolview web (www.evolgenius.info/evolview) (Subramanian et al. Citation2019). The result showed that C. oreastrum is more closely related to C. morifolium in the evolutionary relationship (). This study can be used to provide basic data of the family Asteraceae species for the development of Traditional Chinese Medicine in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. [accessed 2020 Feb 17]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Song C, Liu YF, Song AP, Dong GQ, Zhao HB, Sun W, Ramakrishnan S, Wang Y, Wang SB, Li TZ, et al. 2018. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Mol Plant. 11(12):1482–1491.
- Subramanian B, Gao S, Lercher MJ, Hu S, Chen W-H. 2019. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47(W1):W270–W275.
- Xu YJ, Gao S, Yang YJ, Huang MY, Cheng LN, Wei Q, Fei ZJ, Gao JP, Hong B. 2013. Transcriptome sequencing and whole genome expression profiling of Chrysanthemum under dehydration stress. BMC Genom. 14(1):662.