Abstract
Aristolochia kwangsiensis belongs to Aristolochiaceae family that has many genotoxic plants. Here, we sequenced the complete chloroplast genome of A. kwangsiensis. The genome is 159,849 bp in size with a pair of inverted repeats of 25,570 bp, a large single-copy region of 70,457 bp, and a small single copy region of 19,315 bp. The genome encodes 106 unique genes, including 76 protein-coding, three rRNA, and 27 tRNA genes. Phylogenomic analysis shows that A. kwangsiensis and A. kunmingensis and A. moupinensis form a sister clade. These results will help further studies on taxonomy, phylogeny and evolution of Aristolochiaceae family.
Aristolochia kwangsiensis has been used to clear heat, detoxicate, and stop bleeding in traditional Chinese medicines (Zhou et al. Citation1981; Hoi et al. Citation2019). However, in recent decades, aristolochic acids and their derivatives, chemicals widely contained in Aristolochiaceae plants, are considered genotoxic (Yang et al. Citation2011; Stegelmeier et al. Citation2015; Ng et al. Citation2017). The contradiction between traditional and modern medical views undoubtedly demands in-depth studies of Aristolochiaceae plants. Here, molecular biology research at chloroplast genome level will help us better understand the phylogeny and evolution of this plant family.
Fresh leaf samples were collected from the Guangxi Botanical Garden of Medicinal Plants, Nanning, Guangxi China (E22°85′82″, N108°37′20″). The original plant was identified as A. kwangsiensis by Professor Jinwen You. The genomic DNA was extracted using Plant Genomic DNA kit (Tiangen Biotech, Beijing, China). Genome sequence was performed on the Hiseq 2500 platform (Illumina, San Diego, CA, USA). The voucher specimens and its DNAs (accession number: 201910109) were deposited in the herbarium of the Institute of Medicinal Plant Development at the Chinese Academy of Medical Sciences in Beijing, China. The raw data were assembled into a complete chloroplast genome by using NOVOPlasty (version 2.7.2) (Dierckxsens et al. Citation2017). Annotation was performed using CpGAVAS2 (Shi et al. Citation2019).
The chloroplast genome of A. kwangsiensis (GenBank accession number: MN965793) is 159,849 bp in size with a pair of inverted repeats (IRs) of 25,570 bp separated by a large single-copy (LSC) region of 70,457 bp and a small single-copy (SSC) region of 19,315 bp. The chloroplast genome included 125 genes, of which 106 are unique genes including 76 protein-coding genes, three ribosome RNA (rRNA), and 27 transfer RNA (tRNA) genes. Seven protein-coding genes had one intron and two had two introns.
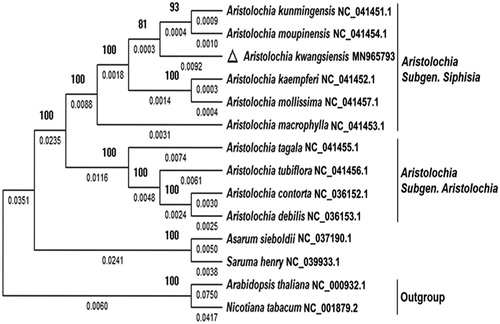
To explore the phylogenomic relationship of Aristolochiaceae, fourteen complete chloroplast genome sequences were downloaded including two outgroup plants. 74 shared proteins were determined and used to reconstruct a maximum-likelihood phylogenetic tree using MEGA-X (version 10.0.4) (Kumar et al. Citation2018). Shared proteins were subjected to multiple sequence alignment using ClustalW module. The evolutionary history was then inferred by Maximum-Likelihood method and JTT matrix-based model. As shown in phylogenetic tree (), 10 species from Aristolochia were clustered together; then, species from Subgenus Siphisia and Subgenus Aristolochia were separated. Aristolochia kwangsiensis and the ancestor of A. kunmingensis and A. moupinensis form a sister clade. The complete chloroplast genome sequence of A. kunmingensis is valuable resources for genome evolution and taxonomy research of Aristolochiaceae.
Figure 1. A phylogenetic tree showing the phylogenetic position of Aristolochia kwangsiensis in Aristolochiaceae based on 74 chloroplast protein sequences constructed with the Maximum-likelihood method. Numerical value below each line shows the branch length; numerical value beside each node shows the bootstrap value obtained from 1000 replications. The GenBank accession number for the corresponding sequences is shown to the right of the Latin name.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):9.
- Hoi TM, Dai DN, Ha CTT, Anh HV, Ogunwande IA. 2019. Essential oil constituents from the leaves of Anoectochilus setaceus, Codonopsis javanica and Aristolochia kwangsiensis from Vietnam. RecNatProd. 13(3):281–286.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Ng AWT, Poon SL, Huang MN, Lim JQ, Boot A, Yu W, Suzuki Y, Thangaraju S, Ng CCY, Tan P, et al. 2017. Aristolochic acids and their derivatives are widely implicated in liver cancers in Taiwan and throughout Asia. Sci Transl Med. 9(412):eaan6446.
- Shi L.C, Chen H.M, Jiang M, Wang L.Q, Wu X, Huang L.F, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stegelmeier B.L, Brown A.W, Welch K.D. 2015. Safety concerns of herbal products and traditional Chinese herbal medicines: dehydropyrrolizidine alkaloids and aristolochic acid. J Appl Toxicol. 35(12):1433–1437.
- Yang H.Y, Wang J.D, Lo T.C, Chen P.C. 2011. Occupational kidney disease among Chinese herbalists exposed to herbs containing aristolochic acids. Occup Environ Med. 68(4):286–290.
- Zhou F.X, Liang P.Y, Qu C.J, Wen J. 1981. Studies on the chemical constituents of Aristolochia kwangsiensis Chun et How ex C F Liang. Yao Xue Xue Bao = Acta Pharmaceutica Sinica. 16(8):638–640.
