Abstract
We first reported the mitochondrial genome of Centropus bengalensis. The mitogenome of C. bengalensis contains 17,117 base pairs. The overall base composition of complete mitogenome is 28.15% A, 27.95% T, 21.86% C, and 22.04% G, with 43.90% of the GC content. All genes exhibit the typical mitochondrial gene arrangement and transcribing directions. Phylogenetic analysis of 4 Centropus species was performed based on the sequence of cytochrome b gene using the neighbor-joining method and the Kimura 2-parameter model in MEGA 7.0.
Centropus bengalensis (Lesser Coucal) majorly distributes in the south of the Yangtze River in China. Meanwhile, it is widely found in South, East, and Southeast Asia. Because the local people are used to killing C. bengalensis as raw material of chicken wine, the number of wild population of the bird has decreased dramatically, and it has been listed as an endangered bird since 1989 by the Chinese government. Compared with other protected species, C. bengalensis received less attention. Hence, less information on the complete mitogenome of C. bengalensis was available. In this study, we characterized the complete mitogenome of C. bengalensis by using next-generation sequencing techniques.
Tissue samples of C. bengalensis were collected from Lai An County (118°43′N, 32°45′E) in Chuzhou City, Anhui Province, in September 2019 and after sampling, the specimens (NJFPC-2019459) were stored in the animal specimens museum of Nanjing Forest-police College.
mtDNA was isolated (Chen et al. Citation2011), 1 μg of purified mtDNA was fragmented and used to construct short-insert libraries (insert size 430 bp) according to the manufacturer’s instructions (Illumina), and then sequenced on the Illumina Hiseq 4000 (Borgstrom et al. Citation2011). Prior to assembly, raw reads were filtered firstly. Then, the filtered reads were assembled into contigs using SOAPdenovo2.04 (Luo et al. Citation2012). The mitochondria genes were annotated using homology alignments and denovo prediction, and the EVidenceModeler v1.1.1 (Haas et al. Citation2008) was used to integrate gene set. A whole mitochondria genome Blast (Altschul et al. Citation1990) search was performed against five databases.
The length of complete mitogenome of C. bengalensis is 17,117 bp (GenBank accession: MN996304). The complete mitogenome is relatively AT rich (that is 56.10% vs 43.90% of the GC content) with the following nucleotide compositions: 28.15% A, 27.95% T, 21.86% C, and 22.04% G. The mitogenome consists of 22 transfer RNA genes, 13 protein-coding genes, two ribosomal RNA genes (i.e., one rrnL and one rrnS), and one control regions. Most of the genes are encoded on the heavy (H) strand, except for those encoding nad6 and eight tRNAs. All genes follow the typical mitochondrial gene arrangement and transcribing directions, which is identical to most vertebrates (Huang et al. Citation2019; Lu et al. Citation2019).
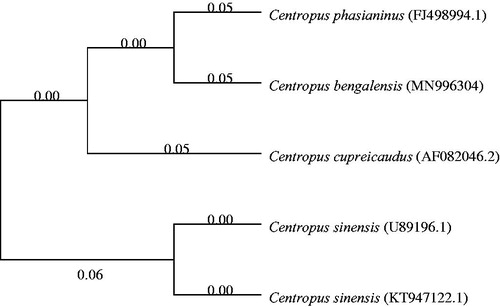
Phylogenetic analysis of 4 Centropus species was performed based on the complete mitogenome of C. bengalensis and the nucleotide sequences of cytochrome b (Cyt b) of other 3 Centropus species, using the neighbour-joining method and the Kimura 2-parameter model in MEGA 7.0, with 1000 bootstrap replicates (Kumar et al. Citation2016). Phylogenetic tree showed that the C. bengalensis is closely related to Centropus phasianinus (). The genome information obtained here could contribute to future studies on molecular evolution and wildlife protection in C. bengalensis.
Disclosure statement
The authors report no conflicts of interest. The sequence has been submitted to NCBI under the accession no. MN996304.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Borgstrom E, Lundin S, Lundeberg J. 2011. Large scale library generation for high throughput sequencing. PLoS One. 6:e19119.
- Chen JM, Guan RZ, Chang SX, Du TQ, Zhang HS, Xing H. 2011. Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L. PLoS One. 6(3):e17662.
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. 2008. Automated eukaryotic gene structure annotation using evidence modeler and the program to assemble spliced alignments. Genome Biol. 9(1):R7.
- Huang YL, Chen YX, Guo HT, Xu YH, Liu HY, Liu DW. 2019. The complete mitochondrial genome sequence of Yarkand hare (Lepus yarkandensis). Mitochondrial DNA B. 4(2):3727–3728.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lu CH, Sun CH, Hou SL, Huang YL, Lu CH. 2019. The complete mitochondrial genome of dark-sided flycatcher Muscicapa sibirica (Passeriformes: Muscicapidae). Mitochondrial DNA B. 4(2):2675–2676.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18–23.