Abstract
The blood-sucking tick Haemaphysalis hystricis is a common ectoparasite of the giant panda and represents a significant threat to both wild and captive populations. Herein, the complete mitogenome of H. hystricis was sequenced using Illumina sequencing technology. The complete mitogenome sequence was 14,715 bp in size and encoded 37 genes including 13 protein-coding genes, 22 transfer RNAs, and two ribosomal RNAs. Phylogeny revealed that two isolates of H. hystricis, regardless of host origins and locations, grouped together and had a closer relationship with Haemaphysalis longicornis than other tick species among the genus Haemaphysalis. The cumulative mitochondrial DNA data provides novel resources for genetic and phylogenetic studies of Haemaphysalis ticks.
The giant panda, Ailuropoda melanoleuca, is regarded as the flagship species for wildlife conservation in China (O’Brien et al. Citation1994; Wang et al. Citation2018). The blood-sucking ticks are common ectoparasites found in giant pandas and can cause dermatitis, anemia, and even death in highly infested wild and captive populations (Qiu and Mainka Citation1993; Cheng et al. Citation2013; Wang et al. Citation2018). About 13 tick species belonging to three genera, namely Haemaphysalis, Ixodes and Dermacentor, have been identified from the giant panda based on morphological studies so far (Wang et al. Citation2018). However, current knowledge progress linked these ticks is limited in their morphology and biology, there are still major gaps in the understanding of the ectoparasites at the molecular level, especially in genetics and molecular epidemiology owing to lacking suitable genetic markers (Cheng et al. Citation2013; Wang et al. Citation2018). Mitochondrial DNA has proven to be a valuable source of molecular markers and is being widely applied for genetics and molecular identification of many zoonotic ectoparasites including ticks (Hwang et al. Citation2001; Cheng et al. Citation2013; Liu et al. Citation2013; Burger et al. Citation2014). In this study, we determined the complete mitogenome sequence of a tick representative Haemaphysalis hystricis from the giant panda in Sichuan Province of China and aimed to provide novel mitochondrial resources to this ectoparasite.
The tick samples (n = 2) were collected from a naturally infected adult male giant panda housed in the Dujiangyan Base of the China Conservation and Research Center for the Giant Panda, Sichuan Province of Southwest China (30°59′N, 103°37′E). Two ticks were identified as H. hystricis females according to the morphological key of Tanskul and Inlao (Citation1989) and the molecular sequencing of the mitochondrial 16S ribosomal DNA gene (Takano et al. Citation2014). One tick specimen was used for DNA extraction and another was archived in the Parasitological Museum of Sichuan Agricultural University (Sichuan, China) under collection numbers XY2018_14. The mitogenome was sequenced using the Illumina HiSeq platform (Novogene, Tianjin, China), and the genome assembly and annotation were performed as previously described (Xie et al. Citation2019). The complete sequence has been deposited in GenBank under accession number: MT013253.
The mitogenome sequence of H. hytricis was 14,715 bp in size with 77.3% AT and encoded 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes. Among the 37 genes, seven PCGs and 14 tRNAs were located on the forward strand (H-strand), whereas the remaining genes were transcribed on the reverse strand (L-strand). Thirteen PCGs, except for nad4 and nad6 deduced to use an incomplete stop codon ‘T’, were predicted to use the typical TAA or TAG as the stop codons. Twenty-two tRNA genes ranged from 51 bp (tRNA(AGA)-Ser) to 68 bp (tRNA-Lys) in size and had a typical clover-leaf like secondary structure. The sizes of two rRNA genes were 696 bp (12S) and 1204 bp (16S), respectively, and present between tRNA-Leu and tRNA-Ile with a separation by tRNA-Val. The control region (also known as D-loop region) was located between tRNA-Leu and tRNA-Cys, similar to other tick species, suggesting its conservation and function in regulation of transcription and control of DNA replication (Clayton Citation1991).
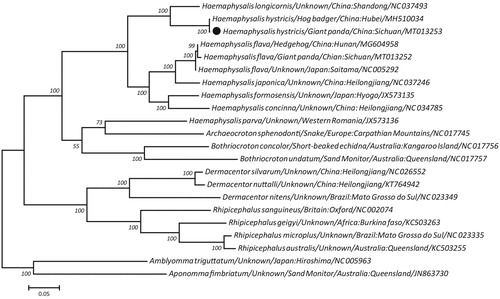
Building on a concatenated amino acid dataset of 12 protein-coding genes from H. hytricis and 20 other ticks, the maximum-likelihood (ML)-based phylogeny demonstrated that two isolates of H. hytricis, regardless of host origins and locations, clustered together and were more closely related to Haemaphysalis longicornis than to other ticks in the genus Haemaphysalis, with 100% bootstrap confidence (), supporting their species validity among the family Ixodidae. In addition, the sister genera including Archaeocroton, Bothriocroton, Dermacentor, and Rhipicephalus in this topology were treated as monophyletic relationships with Haemaphysalis in Ixodidae, consistent with recent molecular studies (Burger et al. Citation2013; Geng et al. Citation2017; Tian et al. Citation2019). Taken together, the H. hytricis mitogenome data sequenced here provides a novel data resource for genetic and evolutionary biological studies of Haemaphysalis ticks.
Figure 1. Maximum-likelihood tree inferred from concatenated amino-acid sequences of twenty mitochondrial PCGs of H. hytricis and other related ticks, utilizing MtArt + G model and 100,000 bootstrap replications. Both Amblyomma triguttatum and Aponomma fimbriatum were used as the outgroups. Values lower than 50% are not shown. The black cycle represents the species in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Burger TD, Shao R, Barker SC. 2013. Phylogenetic analysis of the mitochondrial genomes and nuclear rRNA genes of ticks reveals a deep phylogenetic structure within the genus Haemaphysalis and further elucidates the polyphyly of the genus Amblyomma with respect to Amblyomma sphenodonti and Amblyomma elaphense. Ticks Tick-Borne Dis. 4:265–274.
- Burger TD, Shao R, Barker SC. 2014. Phylogenetic analysis of mitochondrial genome sequences indicates that the cattle tick, Rhipicephalus (Boophilus) microplus, contains a cryptic species. Mol Phylogenet Evol. 76:241–253.
- Cheng WY, Zhao GH, Jia YQ, Bian QQ, Du SZ, Fang YQ, Qi MZ, Yu SK. 2013. Characterization of Haemaphysalis flava (Acari: Ixodidae) from Qingling subspecies of giant panda (Ailuropoda melanoleuca qinlingensis) in Qinling mountains (Central China) by morphology and molecular markers. PLOS One. 8(7):e69793.
- Clayton DA. 1991. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol. 7(1):453–478.
- Hwang UW, Park CJ, Yong TS, Kim W. 2001. One-step PCR amplification of complete arthropod mitochondrial genomes. Mol Phylogenet Evol. 19(3):345–352.
- Geng J, Zheng A, Zou Z, Zhang X. 2017. The complete mitochondrial genome and phylogenetic analysis of Haemaphysalis longicornis Neumann (Acari: Ixodidae). Mitochondrial DNA B Resour. 2(2):856–857.
- Takano A, Fujita H, Kadosaka T, Takahashi M, Yamauchi T, Ishiguro F, Takada N, Yano Y, Oikawa Y, Honda T, et al. 2014. Construction of a DNA database for ticks collected in Japan: application of molecular identification based on the mitochondrial 16S rDNA gene. Jap J Sanit Zool. 65(1):13–21.
- Tanskul P, Inlao I. 1989. Keys to the adult ticks of Haemaphysalis Koch, 1844, in Thailand with notes on changes in taxonomy (Acari: Ixodoidea: Ixodidae). J Med Entomol. 26(6):573–600.
- Tian J, Ge M, Xu H, Wu T, Yu B, Lei C. 2019. The complete mitochondrial genome and phylogenetic analysis of Haemaphysalis hystricis (Parasitiformes: Ixodidae). Mitochondrial DNA B Resour. 4(1):1049–1050.
- Liu GH, Chen F, Chen YZ, Song HQ, Lin RQ, Zhou DH, Zhu XQ. 2013. Complete mitochondrial genome sequence data provides genetic evidence that the brown dog tick Rhipicephalus sanguineus (Acari: Ixodidae) represents a species complex. Int J Biol Sci. 9(4):361–369.
- O’Brien S J, Pan W, Lu Z. 1994. Pandas, people and policy. Nature. 369(6477):179–180.
- Qiu X, Mainka SA. 1993. Review of mortality of the giant panda (Ailuropoda melanoleuca). J Zoo Wildl Med. 24:25–429.
- Wang T, Xie Y, Zheng Y, Wang C, Li D, Koehler Anson V, Gasser RB. 2018. Parasites of the giant panda: a risk factor in the conservation of a species. Adv Parasitol. 99:1–33.
- Xie Y, Liu Y, Gu X, Meng X, Wang L, Li Y, Zhou X, Zheng Y, Zuo Z, Yang G. 2019. Complete mitogenome of the dog cucumber tapeworm Dipylidium caninum (Cestoda, Dilepididae) from Southwest China. Mitochondrial DNA B Resour. 4(2):2670–2672.
