Abstract
As part of a genome sequencing project for Artemisia lavandulaefolia YC (A. lavandulaefolia YC), a complete chloroplast (chl)genome was assembled as a single circular dsDNA of 151,199 bp. It is composed of a large single-copy (LSC), a small single-copy (SSC) and two inverted repeat (IR) regions of 82,909 bp, 18,390 bp and 24,950 bp, respectively. Overall GC contents of the genome were 37.5%. The A. lavandulaefolia YC chloroplast genome has a total of 117 genes including 82 protein-coding genes, 31 tRNA genes, and 4 rRNA genes. Phylogenetic analysis based on the chloroplast genome demonstrated that A. lavandulaefolia YC most closely related to Artemisia motana. The complete chloroplast genomes of A. lavandulaefolia YC would be useful for future investigation of genetics, evolution and clinical identification of artemisia species.
Artemisia lavandulaefolia YC is a perennial herb as a member of the Artemisia belonged to the family Asteraceae (Taleghani et al. Citation2020) and widely distributed in China, Mongolia, Japan and Korea. Artemisia lavandulaefolia YC is mostly born on road side, forest edge, hill-side, shrub and riverside grassland and mentioned as ‘wormwood’ in Europe. Artemisia lavandulaefolia YC, referred to as ‘Ye Aihao’ in China, has been used for the treatment of dispelling cold, dampness and stopping bleeding. Despite its useful applications, there has been little report of molecular and genomic resources (Meng et al. Citation2019). In this article, the chloroplast genome sequence of medicinal plant, A. lavandulaefolia YC, was completely characterized.
The plant samples of A. lavandulaefolia YC were collected from Yangchun of Guangdong province (Latitude: 22°8′54.33″N, Longitude: 111° 67′99.41″E), China and 100% same to KF454262.1 in GenBank by ITS sequence identification. A voucher specimen is deposited at The state Key Laboratory of Plant Physiology and Biochemistry of China with accession number AI_0003. Whole genome sequencing was performed using Illumina genome analyzer (Novaseq, Illumina, San Diego, CA) platform. De novo assembly was performed using clc genome assembler (v.beta 6.0 clc lnc.Aarhus Denmark). We obtained three contigs from de novo genome assembly. Main contigs cover the entire chloroplast genome of reported A. annua chloroplast sequence (Shen et al. Citation2017). Three contigs could rejoined as one single circular complete sequence by manual editing. Gene annotation was conducted using Geseq (Tillich et al. Citation2017) and manual curation through comparison with published chloroplast genomes deposited on NCBI. Inverted repeats were identified using pSTR Finder (Lee et al. Citation2015).The whole genome sequence data reported in this paper have been deposited in the Genome Warehouse in National Genomics Data Center (BIG Data Center Members Citation2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number GWHABJW00000000 that is publicly accessible at https://bigd.big.ac.cn/gwh.
The complete chloroplast genome of A. lavandulaefolia YC is 151,199 bp in size, which is composed of a large single-copy (lsc), a small single-copy (ssc), and two inverted repeat(IR) regions of 82,909 bp, 18,390 bp and 24,950 bp, respectively. The A. lavandulaefolia YC chloroplast genome has a total of 117 genes including 82 protein-coding genes, 31tRNA genes, and 4 rRNA genes.
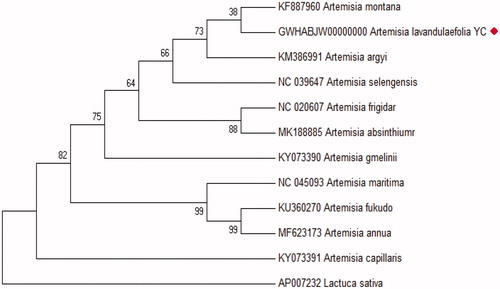
The phylogeny of A. lavandulaefolia YC to other species in the Artemisia (family compositate)was generated. A phylogenetic tree of A. lavandulaefolia YC was constructed using 39 chloroplast protein-coding sequences of A.lavandulaefolia YC with 11 published species in family Artemisia by a maximum likelihood(ML) analysis of MEGA 7.0 (Kumar et al. Citation2016). The phylogenetic tree indicated that A. lavandulaefolia YC was most closely related to A. montana in the genus Artemisia ().
Figure 1. ML phylogenetic tree of Artemisia lavandulaefolia YC with related 11 species of the family Artemisia based on 39 chloroplast protein-coding genes using Mega 7.0 with ML method. Numbers at the nodes are bootstrap values from 2000 replicates. Lactuca sativa (AP007232) was set as the outgroup. All other sequences were downloaded from NCBI GenBank.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- BIG Data Center Members. 2019. Database resources of the BIG Data Center in 2019. Nucleic Acids Res. 47:D8–D14.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lee J.C, Tseng B, Ho B.C, Linacre A. 2015. pSTR Finder: a rapid method to discover polymorphic short tandem repeat markers from whole-genome sequences. Investig Genet. 6(1):10.
- Meng D, Xiaomei Z, Wenzhen K, Xu Z. 2019. Detecting useful genetic markers and reconstructing the phylogeny of an important medicinal resource plant, Artemisia selengensis, based on chloroplast genomics. PLOS One. 14(2):e0211340.
- Shen X, Wu M, Liao B, Liu Z, Bai R, Xiao S, Li X, Zhang B, Xu J, Chen S, et al. 2017. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules (Basel, Switzerland). 22(8):1330–1343.
- Taleghani A, Emami S.A, Tayarani-Najaran Z. 2020. Artemisia: a promising plant for the treatment of cancer. Bioorg Med Chem. 28:115180.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones E S, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
