Abstract
The complete chloroplast (cp) genome sequence of Climacium dendroides, determined using Illumina sequencing data, is presented herein. The DNA sequence of C. dendroides is 124,957 bp in length and has an overall CG content of 29.0%, including 82 protein-coding genes, 36 transfer RNA genes, and 8 ribosomal RNA genes. Phylogenetic trees were constructed based on the cp genome sequences of 10 bryophytes downloaded from GenBank and one acquired from this study.
Climacium dendroides is a type of Bryophyta that belongs to the family Climaciaceae. The genus Climacium was characterized by Friedrich Weber & Daniel Matthias Heinrich Mohr (1804) in Naturhistorische Reise durch einen Theil Schwedens. Climacium is a small group of Hypnales, but it is a morphologically distinct genus (tree-mosses) with four species distributed mainly in the northern hemisphere (Jia et al. Citation2011). Climacium consists of four species, namely C. americanum, C. dendroides, C. japonicum and C. kindbergii. However, only three Climacium species, with the exception of C. kindbergii, have been identified in Korean moss flora (Choe Citation1980). Herein, we determined the complete chloroplast (cp) genome sequence of Climacium dendroides using Illumina sequencing data.
In this study, the cp genome sequence of C. dendroides (GenBank accession no.MT006132), as a representative of Climaciaceae, was sequenced and described. To date, the chloroplast genome of Climaciaceae species has not been reported in the literature. Chloroplast genome sequence of C. dendroides will be useful in the evolutionary studies of bryophytes.
Genomic DNA was extracted from a 5 × 5 cm area collected from the natural habitat of Jilmoen wetland, Odasan (37°45′59.6″N; 128°42′17.6″E) on 5 July 2019. The voucher specimen is stored at the Herbarium of Chonbuk National University (JNU Herbarium) in Korea with the accession number PSJ 20190705-1. Illumina paired-reads (2 × 300 bp) were employed for assembling the chloroplast genome of C. dendroides using Novoplasty 2.4 (Dierckxsens et al. Citation2016) and Geneious R8 8.1.9 (Biomatters Ltd., Auckland, New Zealand). Gene annotation was based on Sanionia uncinata using Geneious R8 8.1.9. A maximum-likelihood tree was constructed using MEGA7 (Kumar et al. Citation2016) based on the JTT matrix-based model, and using concatenated sequences of 16 protein-coding genes (accD, atpA, atpB, cemA, chlB, chlL, clpP, ndhB, ndhF, ndhH, ndhK, petA, psaA, psbC, rbcL and rps2) from 10 related species (7 Bryophyta, 1 Anthocerotophyta and 2 Marchantiophyta) (Kim et al. Citation2019; Cevallos et al. Citation2019).
The cp genome of C. dendroides was 124,957 bp in length and contained 82 PCGs, 36 transfer RNAs (tRNA), and 8 ribosomal RNAs (rRNA). This cp genome has two inverted repeats, each of 9692 bp, a large single-copy (87,030 bp) and small single-copy (18,543 bp). The total GC content of this cp genome is 29.0%, which is similar to that of S. uncinata (29.3%).
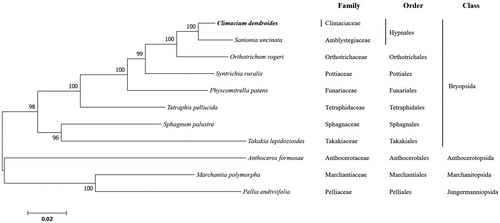
ML analysis showed that C. dendroides first formed a branch with S. uncinata, and then formed a single branch with Hypnales (). The complete chloroplast genome of C. dendroides can provide a reference for developing markers for further studies on the phylogeny and evolution of the Climacium sp. In addition, the cp sequence can be used for species identification or confirmation, which is useful for the conservation and utilization of these multipurpose natural resources.
Figure 1. Phylogenetic position of Climacium dendroides determined by maximum likelihood analysis based on 16 protein coding genes common in all taxa. The bootstrap values above 50% are indicated at each node. Sequences from liverworts were used as outgroup. GenBank accession numbers of mitogenomes used are Anthoceros formosae (NC_004543), Climacium dendroides (MT006132), Marchantia polymorpha (NC_037507), Orthotrichum rogeri (NC_026212), Pellia endiviifolia (NC_019628), Physcomitrella patens (NC_037465), Sphagnum palustre (NC_03019), Syntrichia filaris (MK852705), Takakia lepidozioides (NC_028738) and Tetraphis pellucida (NC_024291).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Cevallos MA, Guerrero G, Ríos S, Arroyo A, Villalobos MA, Porta H. 2019. The chloroplast genome of the desiccation-tolerant moss Pseudocrossidium replicatum (Taylor) RH Zander. Genet Mol Biol. 42(2):488–493.
- Choe DM. 1980. Illustrated flora and fauna of Korea. Vol. 24: Musci. Hepaticae. Seoul: Ministry of Education.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Jia Y, He S, Guo S-L. 2011. Orthotrichaceae. In: Wu P-Ch, Crosby MR, He S, editors. Moss flora of China. English version. Volume 5. Erpodiaceae-Climaciaceae. Beijing, New York: Science Press; St. Louis: Missouri Botanical Garden Press, p. 22–117.
- Kim SC, Byun MY, Kim JH, Lee H. 2019. The complete chloroplast genome of an Antarctic moss Syntrichia filaris (Müll. Hal.). RH Zander. Mitochondrial DNA Part B. 4(2):2303–2304.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
