Abstract
Chloranthus spicatus is an important medicinal and horticultural plant in China. In this study, the complete chloroplastid genome was assembled and annotated, its full-length of 158,758 bp, include large single-copy (LSC) region of 88,408 bp, small single-copy (SSC) region of 18,444 bp, a pair of invert repeats (IR) regions of 26,133 bp. Plastid genome contains 130 genes, 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The phylogenetic analyses based on the complete chloroplast genome sequence provided solid evidence that C. spicatus has a close relationship with C. erectus and C. japonicus. The chloroplast genome presented here will provide useful information for the phylogenetic and evolutionary studies of Chloranthus species.
Chloranthus spicatus (Thunb.) Makino is an evergreen shrub of the chloranthaceae, it is an important medicinal and horticultural plant, which is distributed in Yunnan, Sichuan, Guizhou, Fujian, and Guangdong in China and is widely cultivated in eastern Asia (Xia and Jérémie 1999). Chloranthus spicatus is cultivated for its aromatic leaves and flowers which are used to make a tea-like drink (Zhang and Ma Citation2005). When it is used as external application of medicinal materials, its roots and stems are chopped into mud, or dried and ground into fine powder, which can be used for the treatment of fractures, dislocations, sprains and bruises, periarthritis of shoulder, protrusion of lumbar disc, and so on (Cao et al. Citation2008). The classification status of this species is controversial according on the datum. Therefore, we assembled and annotated the complete chloroplast genome of C. spicatus. The genomic data will be useful for population and phylogenetic studies of C. spicatus.
Fresh leaf sample of C. spicatus was acquired from Taining (26°54′42.5″N, 117°05′38.90″E), Fujian province of China, and voucher specimen deposited at Herbarium of College of Forestry, Fujian Agriculture and Forestry University (specimen code Liao Csp01). DNA extraction from fresh leaf tissue, with 350 bp randomly interrupted by the Covaris ultrasonic breaker for library construction. The constructed library was sequenced using PE150 Illumina Hiseq Xten platform, approximately 2GB data were generated. Illumina data were filtered by script in the cluster (default parameter: -L 5, -p 0.5, -N 0.1). Plastid genome was assembled using GetOrganelle pipe-line (https://github.com/Kinggerm/GetOrganelle), it can get the plastid-like reads, and the reads were viewed and edited by Bandage (Wick et al. Citation2015). Assembled chloroplast genome annotation based on comparison with C. spicatus by GENEIOUS R11.15 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. Citation2012). The annotation result was drawn with the online tool OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013).
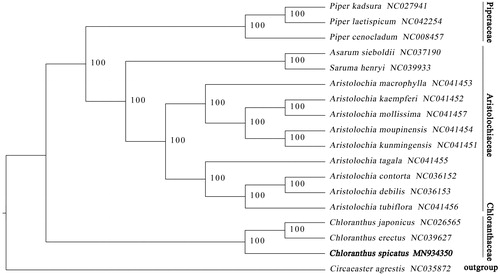
The complete plastid genome sequence of C. spicatus (GenBank accession MN 934350) was 158,758 bp in length, with a large single-copy (LSC) region of 88,408 bp, a small single-copy (SSC) region of 18,444 bp, and a pair of inverted repeats (IR) regions of 26,133 bp. Complete chloroplastid genome contains 130 genes, there were 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The complete genome GC content was 39.20%. To study the phylogenetic relationship of C. spicatus with other members of Chloranthaceae, 16 species (Aristolochia contorta, Aristolochia debilis, Aristolochia kaempferi, Aristolochia kunmingensis, Aristolochia macrophylla, Aristolochia mollissima, Aristolochia moupinensis, Aristolochia tagala, Aristolochia tubiflora, Asarum sieboldii, Saruma henryi, Piper cenocladum, Piper kadsura, Piper laetispicum, Chloranthus erectus, and Chloranthus japonicus) of Aristolochiaceae, Piperaceae, and Chloranthaceae, Circaeaster agrestis from Circaeasteraceae as outgroup, all of them downloaded from NCBI GenBank. The sequences were aligned using MAFFT v7.388 (Katoh and Standley Citation2013), and phylogenetic tree constructed using RAxML (Stamatakis Citation2014) (). The relationship between Chloranthus and Circaeaster was strongly supported (bootstrap = 100%). The maximum-likelihood phylogenetic analysis shows that C. spicatus was closely related to C. erectus and C. japonicus.
Acknowledgements
We cordially thank Di-Yang Zhang and Bin Liu for their kind help during the preparation of this paper.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cao CM, Peng Y, Xiao PG. 2008. Advance in on chemical constituent and bioactivity research of genus Chloranthus. CJCMM. 33(13):7–13.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Xia N, Jérémie J. 1999. Chloranthaceae. In: Editorial Committee, editor. Flora of China. Vol. 4. Beijing: Science Press; p. 133–138.
- Zhang LX, Ma J. 2005. Cultivation and utilization of Chloranthus. China Trop Agric. (4):32–33.