Abstract
Parantica aglea (Lepidoptera: Nymphalidae: Danainae) was the type species of the genus Parantica. In this study, the complete mitochondrial genome (mitogenome) of P. aglea (GenBank accession No. MN938921) was sequenced using the polymerase chain reaction (PCR) method. Its mitogenome was a circular molecule of 15,214 bp in length, containing 13 protein-coding genes, two ribosomal RNAs, 22 transfer RNAs, and one AT-rich region. The A + T content of the overall base composition of H-strand was 79.6%. All of the 13 protein-coding genes were begun with typical start codon ATN except COI using CGA. Four genes (COI, COII, ND4, and ND5) were ended with an incomplete stop codon (TA– or T––), and the remaining nine terminated with TAN. In the Bayesian inference (BI) phylogenetic tree, P. aglea was positioned near P. sita niphonica and P. s. sita within the genus Parantica.
The glassy tiger butterfly, Parantica aglea (Lepidoptera: Nymphalidae: Danainae), as the type species of the genus Parantica, mainly distributes in South Asia, East Asia, Southeast Asia, and Papua New Guinea of Oceania (Zhang et al. Citation2008). In the present study, we sequenced the complete mitochondrial genome of the P. aglea (GenBank accession No. MN938921), representing the new record of complete mitochondrial genome from the genus Parantica.
Adult specimens of P. aglea were collected in Yizhou in Guangxi Province, China. The collected specimens were stored in 95% ethanol at temperature −20 °C and deposited in the Museum of Insects of Hechi University (the voucher No. L387), Yizhou, Guangxi. Whole genomic DNA was extracted from the abdomen of each specimen using a Wizard® Genomic DNA Purification Kit (Promega, Madison, USA) according to the manufacturer’s instructions. The mitogenome of Parantica sita sita (GenBank accession No. MG571524) was employed as the reference sequence (Hu and Wang Citation2019). Certain pairs of universal primers for butterfly mitochondrial genomes were used for polymerase chain reaction (PCR) amplification (Simon et al. Citation2006). Then, PCR products were sequenced using primer-walking strategy. The complete mitochondrial genome was assembled by SeqMan program from DNASTAR (Burland Citation2000) and annotated using MITOS Web Server (Bernt et al. Citation2013).
We obtained complete mitogenome of P. aglea with 15,214 bp long, which consisted of 13 typical protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one AT-rich region, which is similar to the typical mtDNA of other insects (Li et al. Citation2019). Among the mitochondrial protein-coding genes, ATP8 gene was the shortest, whereas ND5 the longest. Twelve PCGs started with typical ATN codon (one with ATC, two with ATA, three with ATT, six with ATG), whereas COI appeared to start with CGA. Nine PCGs terminated with TAN codon (seven with TAA and two with TAG), and the remaining four terminated with an incomplete stop codon TA– or T––. The 22 tRNA genes ranged in size from 60 to 71 bp. The 12S rRNA (757 bp) and 16S rRNA (1368 bp) were located between the trnL1 and AT-rigion and separated by the trnV gene. The AT-rich region of the mitogenome was 472 bp long and located between the trnV gene and trnM gene. The overall base composition of the entire genome was as follows: A (38.4%), T (41.2%), C (12.7%), and G (7.7%), reflected a typical sequence feature of the insect mitogenome.
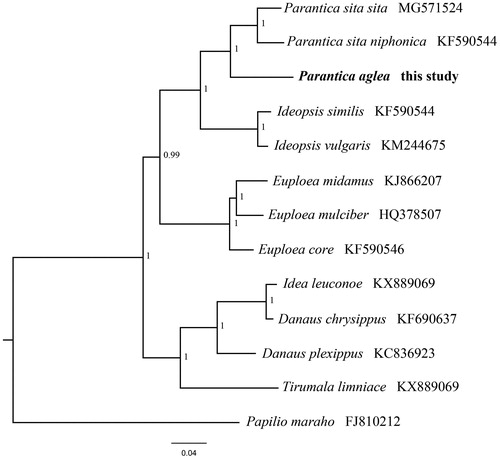
To validate the phylogenetic position of P. aglea, the Bayesian inference (BI) tree was constructed on CIPRES Portal using 13 mitochondrial PCGs from mitogenomes of 12 species and one outgroup from Papilionidae, respectively. We used the best-fit partitioning scheme and partition-specific models recommended by PartitionFinder (Lanfear et al. Citation2012). As shown in , P. aglea was positioned near Parantica sita niphonica and P. s. sita within the genus Parantica. The result of phylogenetic analysis was consistent with the previous research (Hu and Wang Citation2019). It indicated that our newly determined mitogenome sequence could meet the demands and explain some evolution issues.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Burland TG. 2000. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 132:71–91.
- Hu P, Wang R. 2019. The complete mitochondrial genome of Parantica sita sita (Lepidoptera: Nymphalidae: Danainae) revealing substantial genetic divergence from its sibling subspecies P. s. niphonica. Gene. 686:76–84.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Li R, Shu X, Li X, Meng L, Li B. 2019. Comparative mitogenome analysis of three species and monophyletic inference of Catantopinae (Orthoptera: Acridoidea). Genomics. 11(6):1728–1735.
- Simon C, Buckley T, Frati F, Stewart J, Beckenbach A. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymeras chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 37(1):545–579.
- Zhang YL, Fang LJ, Chou I. 2008. Taxonomic study on Chinese species of the genus Parantica Moore (Lepidoptera, Nymphalidae, Danainae). Acta Zootaxon Sin. 33(1):157–163.