Abstract
Chlamydomonas moewusii is a microalga isolated from the Tarim Basin of Xinjiang, China. The complete mitochondrial genome sequence of C. moewusii strain XJCH-01 was determined in this study (Accession number MT015649). The mitogenome (22,887 bp, 34.58% G + C) consists of 7 protein-coding genes (PCG), discontinuous large and small subunit ribosomal RNA (rRNA), and 4 transfer RNA (tRNA) genes. The complete mitochondrial genome sequence of the C. moewusii strain XJCH-01 enriches data resources for further study in genetic and functional evolution.
The single-celled Chlamydomonas was similar to the ancestors of land plants, which is considered an interesting model organism for studying genetic and functional evolution (Merchant et al. Citation2007). Chlamydomonas moewusii is a genus of the family Chlamydomonadaceae in the order Chlamydomonadales of Chlorophyceae consisting of unicellular flagellates, found in stagnant water and on damp soil containing organic matter, as a hydrogen-producing ecologically important group in the environment (Harris Citation2001). Here, the complete mitochondrial genome of C. moewusii was sequenced and characterized in detail. The strain of C. moewusii, named as XJCH-01, was isolated from Tarim Basin (39°95′N 84°26′E), Xinjiang Uygur Autonomous region, China in August, 2019. It was stored in the College of Life Sciences, Linyi University, Linyi, China. Genomic DNA was extracted from cultured C. moewusii according to Liu et al. (Citation2012, Citation2014). The complete mitochondrial genome of C. moewusii strain XJCH-01 was sequenced using a shotgun approach and assembly. Subsequently, sequence data were analyzed according to Denovan-Wright et al. (Citation1998). Clustal W was used for multiple sequence alignments (Thompson et al. Citation1994) and nucleotide sequence similarity searches were performed at the National Center for Biotechnology Information using the BLAST online service (Altschul et al. Citation1990).
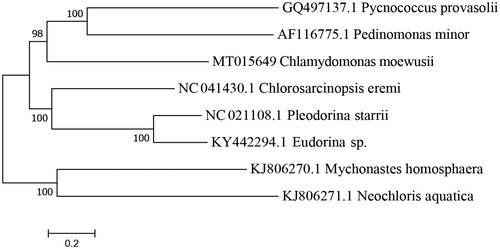
The complete mitochondrial genome C. moewusii strain XJCH-01 (Accession number MT015649) was 22,887 bp in length with a G + C content of 34.58%. The C. moewusii mtDNA encodes seven respiratory chain proteins (cob: apocytochrome b, cox1: subunit 1 of cytochrome oxidase, subunits nad1, nad2, nad4, nad5, and nad6: 1, 2, 4, 5, and 6 of the NADH dehydrogenase complex), four tRNAs and discontinuous LSU and SSU rRNAs. However, there are two large direct repeat regions that do not appear to encode any functional mitochondrial molecule. The arrangement and composition of the mitochondrial genome are similar to the known microalgae genomes (Wolff et al. Citation1994; Denovan-Wright et al. Citation1998; Smith et al., Citation2011; Servin-Garciduenas and Martinez-Romero Citation2012; Tourasse et al. Citation2015; Hu et al. Citation2019; Huang et al. Citation2019). However, it showed a big difference from the vertebrates (Liu et al. Citation2016; Li, Liu, Sui, et al. Citation2019; Li, Kiu, Zhang, Citation2019) and the Arthropoda (Lavrov et al. Citation2000; Masta and Boore Citation2004; Choi et al. Citation2007). A phylogenetic tree was constructed based on the comparison of the complete mitochondrial genome sequences with other Chlorophyceae species using neighbour-joining method to determine the phylogenetic position of C. moewusii strain XJCH-01 (). All 7 protein-coding genes identified in the C. moewusii mitochondrial genome, started with the typical initiation codon ATG. No incomplete termination codons were found, all PCGs had the complete termination codons TAA, whereas cob terminate with TAG. Among them, three (cob, cox1, and nad1) each contain one group I intron and one (nad5) contains two group I introns. In addition, seven ORFs (i1-orf, i2-orf, i3-orf, i4-orf, i5-orf, i6-orf, i7-orf) were identified within the nine introns interrupting the coding regions of C. moewusii mtDNA. There is an additional gene for tRNA-met in C. moewusii mtDNA. Two of the tRNA genes, tRNA-Trp and tRNA-Gln, are clustered and separated by only 7 bp, whereas tRNA-met-1 and tRNA-met-2 were located between the two LSU regions, and separated by a large direct repeat region. The mitochondrial SSU and LSU rRNAs are encoded by three (rns-a ∼ rns-c) and six (rnl-a ∼ rnl-f) rRNA gene pieces, respectively, which were dispersed throughout the genome and interspersed with each other and with protein-coding and tRNA genes. All the mitogenome genes were encoded on the same strand of the mtDNA molecule. The complete mitochondrial genome sequence obtained in our study would facilitate further investigations of phylogenetic relationships, genomic evolution and function within Chlorophyceae.
Figure 1. A phylogenetic tree constructed based on the comparison of complete mitochondrial genome sequences with of the other seven Chlorophyceae species. The seven Chlorophyceae species are Pycnococcus provasolii, Pedinomonas minor, Chlorosarcinopsis eremi, Pleodorina starrii, Iudorina sp., Mychonastges homosphaera, and Neochloris aquatica. Genbank accession numbers for all sequences are listed in the figure. The numbers at the nodes are bootstrap percent probability values based on 1000 replications.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Choi HE, Park SJ, Jang HK, Hwang W. 2007. Complete mitochondrial genome of a Chinese scorpion Mesobuthus martensii (Chelicerata, Scorpiones, Buthidae). DNA Seq. 18(6):461–473.
- Denovan-Wright EM, Nedelcu AM, Lee RW. 1998. Complete sequence of the mitochondrial DNA of Chlamydomonas eugametos. Plant Mol Biol. 36(2):285–295.
- Harris EH. 2001. Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol. 52(1):363–406.
- Hu S, Xu Y, Xu B, Li F. 2019. The complete mitochondrial genome of a marine microalgae Cryptophyceae sp. CCMP2293, Mitochondrial DNA Part B. 4(1):848–849.
- Huang L, Gao B, Wang F, Zhang C. 2019. The complete mitochondrial genome of an oleaginous microalga Vischeria stellata strain SAG 33.83 (Eustigmatophyceae). Mitochondrial DNA Part B. 4(1):301–302.
- Lavrov DV, Boore JL, Brown WM. 2000. The complete mitochondrial DNA sequence of the Horseshoe Crab Limulus polyphemus. Mol Biol Evol. 17(5):813–824.
- Li YY, Liu LX, Sui ZH, Ma C, Liu YG. 2019. The complete mitochondrial DNA sequence of Mongolian grayling Thymallus brevirostris. Mitochondrial DNA Part B. 4(1):1204–1205.
- Li YY, Liu LX, Zhang RH, Ma C, Liu YG. 2019. The complete mitochondrial DNA sequence of ruffe Acerina cernua. Mitochondrial DNA Part B. 4(1):1590–1591.
- Liu YG, Guo YG, Hao J, Liu LX. 2012. Genetic diversity of swimming crab (Portunus trituberculatus) populations from Shandong peninsula as assessed by microsatellite markers. Biochem Syst Ecol. 41:91–97.
- Liu YG, Li YY, Meng W, Liu LX. 2016. The complete mitochondrial DNA sequence of Xinjiang arctic grayling Thymallus arcticus grubei. Mitochondrial DNA Part B. 1(1):724–725.
- Liu YG, Liu LX, Xing SC. 2014. Development and characterization of thirteen polymorphic microsatellite loci in the Cynoglossus semilaevis. Conserv Genet Resour. 6(3):683–684.
- Masta SE, Boore JL. 2004. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol. 21(5):893–902.
- Servin-Garciduenas LE, Martinez-Romero E. 2012. Complete mitochondrial and plastid genomes of the green microalga Trebouxiophyceae sp. Strain MX-AZ01 isolated from a highly acidic geothermal lake. Eukaryot Cell. 11(11):1417–1418.
- Smith DR, Burki F, Yamada T, Grimwood J, Grigoriev IV, Van Etten JL, Keeling PJ. 2011. The GC-rich mitochondrial and plastid genomes of the green alga Coccomyxa give insight into the evolution of organelle DNA nucleotide landscape. PLoS One. 6(8):e23624.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 22(22):4673–4680.
- Tourasse NJ, Shtaida N, Khozin-Goldberg I, Boussiba S, Vallon O. 2015. The complete mitochondrial genome sequence of the green microalga Lobosphaera (Parietochloris) incisa reveals a new type of palindromic repetitive repeat. BMC Genomics. 16(1):580.
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 318(5848):245–250.
- Wolff G, Plante I, Lang BF, Kuck U, Burger G. 1994. Complete sequence of themitochondrial DNA of the chlorophyte alga Prototheca wickerhamii. Gene content and genome organization. J Mol Biol. 237(1):75–86.
