Abstract
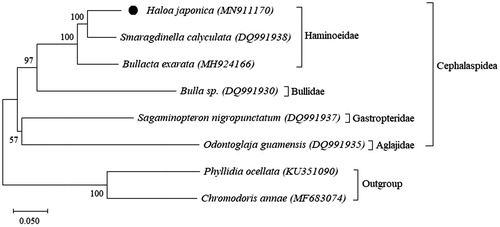
Haloa japonica is a bubble snail species in the family Haminoeidae. In the present study, the complete mitochondrial genome of H. japonica was decoded and analyzed. With the size of 13,797 bp, the mitogenome of H. japonica is the shortest among all known mitogenomes of the order Cephalaspidea. Overall base-pair composition of the mitogenome was 69% A–T and 31% C–G. The phylogenetic tree showed a clade of H. japonica and other species in the family Haminoeidae.
Haloa japonica, previously known as Haminoea japonica (WoRMS Editorial Board Citation2020), is a bubble snail with native distribution in Japan and Korea. Moreover, non-native populations were reported in different regions of the world (Gosliner and Behrens Citation2006). The schistosome parasite found in H. japonica is involved in human cercarial dermatitis (Brant et al. Citation2010). Therefore, the spread of H. japonica could negatively impact human health. However, it has been challenging to detect H. japonica populations in non-native regions due to difficulties in the identification of H. japonica and its related species (Hanson et al. Citation2013). Mitogenome is widely used for resolving phylogenetic and taxonomic complications. The complete mitogenome is necessary for discrimination of H. japonica with its related species.
The sample of H. japonica was collected from Jeju Island, Korea (33°28′59.45″N, 126°22′26.73″E) in May 2013. The sample (voucher number: SMU00051) was deposited in the Department of Biotechnology, Sangmyung University, Korea. The procedure of mitogenome sequencing and annotation for H. japonica was presented in the previous study (Do et al. Citation2019). For phylogenetic analysis, the mitogenomes of cephalaspidean species were retrieved from GenBank. The phylogenetic analysis using amino acid sequences of 13 protein-coding genes (PCGs) was performed by MEGA X (Kumar et al. Citation2018) with the neighbor-joining method. The bootstrap method (×1000 replicate) was used to calculate statistical support.
The total length of the H. japonica mitogenome (GenBank accession number: MN911170) was 13,797 bp, with 13 PCGs, 2 ribosomal RNA genes, and 22 tRNA genes. This is the shortest size among all recorded mitogenomes of the order Cephalaspidea. The main reason of the short length could be the compact structure of the mitogenome. Accordingly, there were up to 28 overlapping regions found in the mitogenome. The base-pair composition of the H. japonica mitogenome included 69% A–T and 31% C–G. Among 37 genes of the mitogenome, there were 24 genes encoded on the H-strand and 13 genes encoded on the L-strand. The gene order of the H. japonica mitogenome is similar to the mitogenomes of Smaragdinella calyculata and Bullacta exarata, which also belong to the family Haminoeidae (Worms Editorial Board Citation2020).
Of 13 PCGs, eight genes initiated with ATG (atp8, atp6, cob, cox3, nd1, nd4, nd5, and nd6) and five remaining genes initiated with ATT (cox1, cox2, nd2, nd3, and nd4l) codons. For stop codon, seven genes terminated with TAA (atp8, atp6, cob, cox1, nd1, nd4l, and nd6), three genes terminated with TAG (cox2, nd2 and nd5) and three genes had incomplete termination with T- (cox3, nd3 and nd4).
The phylogenetic analysis revealed three species in the family Haminoeidae are clustered together, and H. japonica is closest to Smaragdinella calyculata (). This pattern is consistent with the report on the relationship of the family Haminoeidae (Oskars et al. Citation2019). Our study presented the mitogenome of H. japonica and it is the third record for the family Haminoeidae. Mitogenome sequencing for other haminoeid species are recommended to resolve taxonomic difficulties for this group.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Brant SV, Cohen AN, James D, Hui L, Hom A, Loker ES. 2010. Cercarial dermatitis transmitted by exotic marine snail. Emerg Infect Dis. 16(9):1357–1365.
- Do TD, Choi TJ, Jung DW, Kim JI, Karagozlu MZ, Kim CB. 2019. The complete mitochondrial genome of Phyllidiella pustulosa (Cuvier, 1804) (Nudibranchia, Phyllidiidae). Mitochondrial DNA B. 4:771–772.
- Gosliner TM, Behrens BW. 2006. Anatomy of an invasion: systematics and distribution of the introduced opisthobranch snail, Haminoea japonica Pilsbry, 1895 (Gastropoda: Opisthobranchia: Haminoeidae). Proc Calif Acad Sci. 7:1003–1010.
- Hanson D, Cooke S, Hirano Y, Malaquias MAE, Crocetta F, Valdes A. 2013. Slipping through the cracks: the taxonomic impediment conceals the origin and dispersal of Haminoea japonica, an invasive species with impacts to human health. PLOS One. 8(10):e77457.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Oskars TR, Too CC, Rees D, Mikkelsen PM, Willassen E, Malaquias M. 2019. A molecular phylogeny of the gastropod family Haminoeidae sensu lato (Heterobranchia, Cephalaspidea) - a generic revision. Inv Syst. 33:426–472.
- WoRMS Editorial Board. 2020. World register of marine species. [accessed 2020 Jan 04]. http://www.marinespecies.org