Abstract
Protaeolidiella atra is an aeolid nudibranch found in Korean and Japanese waters. This study presents the complete mitochondrial genome of P. atra from Korea. The mitogenome size of P. atra was 14,445 bp, including 27.3% A, 39.3% T, 14.8% C, and 18.6% G. Identical to other nudibranchs, there were 13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes in the mitogenome of P. atra. The result of the phylogenetic analysis indicated that P. atra is closest to Sakuraeolis japonica.
In 1955, Baba described Protaeolidiella atra for the first time based on samples collected from Japan (Baba Citation1955). Even though P. atra and Pleurolidia juliae were once considered to be conspecific, morphological and molecular pieces of evidence proved that they are two distinct species (Carmona et al. Citation2015). Protaeolidiella atra was previously regarded as a primitive species of the family Aeolidiidae. However, the differences in morphology and specialized diet of P. atra compared to other members of the family have resulted in a long debate on the phylogenetic position of this species (Carmona et al. Citation2015). Currently, P. atra and P. juliae are accepted as only two species of the family Pleurolidiidae (WoRMS Editorial Board Citation2020). Decoding and analysis of the P. atra mitogenome might be useful for the understanding of its phylogenetic relationships.
In this study, P. atra was sampled from Ulleungdo island, Korea (37°32′20.40″N, 130°50′19.82″E) in August 2013. The collected sample (voucher number: SMU00041) was preserved in absolute ethanol and maintained in the Department of Biotechnology, Sangmyung University, Korea. Following DNA extraction, Next-generation sequencing was conducted on Miseq system (Illumina, San Diego, CA), and the paired-end reads were assembled by MITObim (Hahn et al. Citation2013). The mitogenome sequence was annotated using the MITOS web server (Bernt et al. Citation2013) with the support of ARWEN (Laslett and Canbäck Citation2008) for tRNA search. The phylogenetic position of P. atra was determined based on amino acid sequences of protein-coding genes (PCGs). The tree of P. atra and related species was reconstructed with the neighbor-joining method in MEGA X (Kumar et al. Citation2018).
The size of the complete mitogenome of P. atra (GenBank accession number: MN911169) was 14,445 bp with nucleotide composition: 27.3% A, 39.3% T, 14.8% C, and 18.6% G. Nine of the 13 PCGs were encoded on the H-strand (cob, cox1, cox2, nd1, nd2, nd4, nd4l, nd5, and nd6) while four remaining PCGs were encoded on the L-strand (atp6, atp8, cox3, and nd3). The sizes of PCGs were in the range of 153 bp (atp8) to 1680 bp (nd5). Of the 22 tRNA genes, the sizes varied from 54 bp (tRNAtyr) to 69 bp (tRNAGly). For rRNA genes, 12S rRNA gene was 721 bp in length and encoded on the L-strand while 16S rRNA gene was 1140 bp in length and encoded on the H-strand.
There were 18 overlapping regions with the longest region found between tRNAIle and nd2 (40 bp). In addition, 18 intergenic regions were detected, ranging from 1 to 240 bp and the longest region was located between tRNAHis and tRNACys.
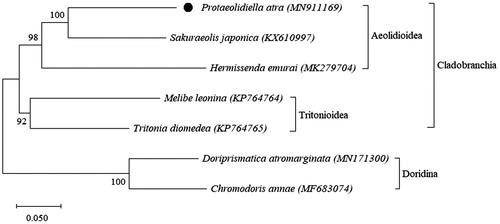
The phylogenetic tree showed the position of P. atra in the suborder Cladobranchia (). Protaeolidiella atra formed a clade with Sakuraeolis japonica, and they were sister to Hermissenda emurai (). This study provided the first mitogenome record for the family Protaeolidiellidae. Additional data from Pleurolidia juliae and different species of the superfamily Aeolidioidea is necessary to reveal insight into the phylogenetic position of P. atra.
Figure 1. Phylogenetic relationships of P. atra and species in the suborder Cladobranchia. The black spot indicates the mitogenome of P. atra. The complete mitogenomes of remaining nudibranchs are obtained from GenBank. Chromodoris annae and Doriprismatica atromarginata (suborder Doridina) represents the outgroup.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Baba K. 1955. Opisthobranchia of Sagami Bay supplement. Iwanami Shoten, Tokyo. 59 pp.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Carmona L, Pola M, Gosliner TM, Cervera L. 2015. Protaeolidiella atra Baba, 1955 versus Pleurolidia juliae Burn, 1966: one or two species? Helgol Mar Res. 69(2):137–145.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- WoRMS Editorial Board. 2020. World Register of Marine Species; [Accessed 2020 Jan 03]. http://www.marinespecies.org.
