Abstract
We determined the complete chloroplast genome sequence of Viola japonica using Illumina pair-end sequencing data. The whole chloroplast genome was 156,504 bp in length, consisting of a pair of inverted repeats (IR) of 26,427 bp, a large single-copy (LSC) region of 85,647 bp, and a small single-copy (SSC) region of 18,003 bp. The gene contents and order were exactly the same as published five Violaceae plastomes. A total of 111 unique genes were annotated, including 77 protein-coding genes, 30 tRNAs, and 4 rRNAs. Among the annotated genes, 16 genes contained one or two introns. The ML tree showed that V. japonica formed a clade with Viola seoulensis.
Violaceae Batch. consists of approximately 22 genera and 1000–1100 species of herbs, shrubs, lianas, and trees (Wahlert et al. Citation2014; Marcussen et al. Citation2015; Cheon et al. Citation2019). Viola L., the largest genus in the Violaceae, comprises 583–620 species and is distributed mainly in temperate and tropical regions (Clausen Citation1967; Yoo and Jang Citation2010; Wahlert et al. Citation2014, Cheon et al. Citation2019). This genus is known as one of the more difficult groups to classify and although many phylogenetic studies carried out, the phylogenetic relationships are still unclear among the section and species (Clausen Citation1967; Ballard et al. Citation1998; Yockteng et al. Citation2003; Yoo et al. Citation2005; Liang and Xing Citation2010; Yoo and Jang Citation2010).
In this study, we report the whole cp genome sequence of V. japonica to gain information for further studies on the phylogenomics. The sampled V. japonica was collected from Minoreum (33°28′32.9′′N, 126°30′29.8′′E), Jeju-si in Korea. The specimen (voucher number KWNU93664) was deposited the Kangwon National University Herbarium (KWNU). Total DNA was extracted using a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA). Genomic DNA was used for sequencing with the Illumina Miseq (Illumina Inc., San Diego, CA) platform. Viola japonica was sequenced to produce 5,862,188 raw reads and 301 bp were obtained from them. Using these sequence data, we assembled the complete chloroplast genome with Geneious 7.1 (Biomatters Ltd, Auckland, New Zealand). Annotation of the chloroplast genome was based on an online available program, DOGMA (Wyman et al. Citation2004), coupled with manual corrections for start and stop codons. We also compared each gene to the published complete chloroplast genome sequences of Campanulaceae for correct gene annotation. The tRNAs were confirmed using tRNAscan-SE (Schattner et al. Citation2005). A circular plastid genome map was drawn using the OGDRAW program (Greiner et al. Citation2019).
The plastid genome of V. japonica (GenBank accession no. MT012304) is a circular DNA molecule 156,504 bp in length with 36.3% G + C content, consisting of a pair of inverted repeats (IR) of 26,427 bp, a large single-copy (LSC) region of 85,647 bp, and a small single-copy (SSC) region of 18,003 bp. The cp genome contains a total of 111 unique genes constituting 77 protein-coding genes, 30 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes. The gene contents and order were exactly the same with published five Violaceae plastomes (Viola mirabilis, Viola phalacrocarpa, Viola raddeana, Viola seoulensis, and Viola websteri). Among the annotated genes, 16 genes contain one or two introns (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, petB, petD, atpF, ndhA, ndhB, clpP, rpl2, rpl16, rps12, rpoC1, and ycf3).
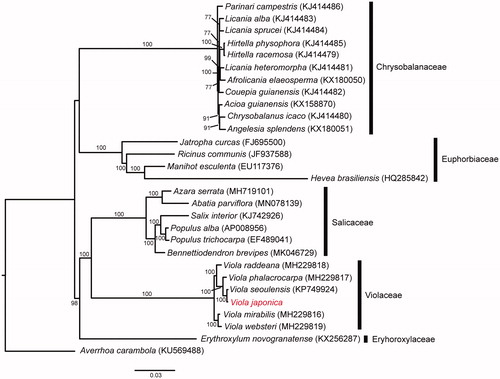
To investigate the phylogenetic relationships among the Malphigiales, we conducted a phylogenetic analysis. A total of 76 protein-coding genes from 28 Malphigiales species and one outgroup (Averrhoa carambola) were aligned using MAFFT (Katoh et al. Citation2002). We then conducted maximum-likelihood (ML) analysis using RAxML v.7.4.2 with 1000 bootstrap replicates and the GTR + I model (Stamatakis Citation2006). The phylogenetic tree () is clearly divided into five clades at the family level. The Violaceae was monophyletic with a high support value (BS = 100) and was divided into two clades. Also, V. japonica was closely related to V. seoulensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ballard HE, Sytsma KJ, Kowal RR. 1998. Shrinking the violets: phylogenetic relationships of infrageneric groups in Viola (Violaceae) based on internal transcribed spacer DNA sequences. Syst Bot. 23(4):439–458.
- Cheon KS, Kim KA, Kwak M, Lee B, Yoo K. 2019. The complete chloroplast genome sequences of four Viola species (Violaceae) and comparative analysis with its congeneric species. PLoS One. 14(3):e0214162.
- Clausen J. 1967. Cytotaxonomy and distributional ecology of western North American violets. Madroño. 17:173–204.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expended toolkit for the graphical visualization of organellar genomes. Nucleic Acid Res. 47(W1):W59–W64.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Liang GX, Xing FW. 2010. Infrageneric phylogeny of the genus Viola (Violaceae) based on trnL-trnF, psbA-trnH, rpl16, ITS sequences, cytological and morphological data. Acta Botanica Yunnanica. 32:477–488.
- Marcussen T, Heier L, Brysting AK, Oxelman B, Jakobsen S. 2015. From gene trees to a dated allopolyploid network: insights from the Angiosperm genus Viola (Violaceae). Syst Biol. 64(1):84–101.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Wahlert GA, Marcussen T, De Paula-Souza J, Feng M, Ballard HE. 2014. A phylogeny of the Violaceae (Malpighiales) inferred from plastid DNA sequences: implications for generic diversity and intrafamilial classification. Syst Bot. 39(1):239–252.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yockteng R, Ballard HE, Mansion G, Dajoz I, Nadot S. 2003. Relationships among pansies (Viola section Melanium) investigated using ITS and ISSR markers. Plant Syst Evol. 241(3–4):153–170.
- Yoo KO, Jang SK. 2010. Infrageneric relationships of Korean Viola based on eight chloroplast markers. J Syst Evol. 48(6):474–481.
- Yoo KO, Jang SK, Lee WT. 2005. Phylogeny of Korean Viola based on ITS sequences. Korean J Plant Taxon. 35(1):7–23.