Abstract
Phascolosoma is one of the most important genera of Sipuncula, many species of which including P. similis are economic important species in the fishery of southeast China. However, the current taxonomy determination of P. similis had not been verified yet. Here, we report the complete mitochondrial genome sequence of P. similis. The mitogenome has 16,237 base pairs and made up of a total of 38 genes (13 protein-coding, 23 transfer RNAs, and 2 ribosomal RNAs), and a putative control region. The complete mitogenomes of P. similis will provide useful genetic information for future phylogenetic and taxonomic classification of Phascolosoma.
Sipuncula (known as peanut worms) is a relatively small taxon which contain about 150 exclusively species with a global distribution (Schulze et al. Citation2012). Phascolosoma is one of the most important genera of Sipuncula, many species of which are geographically widespread and have planktonic pelagosphera larvae. However, the synonym and misidentification of species in Phascolosoma have been reported repeatedly (Cutler and Cutler Citation1983). It had been debated whether or not Phascolosoma similis is a valid taxon of Phascolosoma (Li Citation1989). Mitochondrial genome is an excellent molecular marker for phylogenetic analysis and taxonomy identification, especially when there is a small number of unambiguous taxonomic characters available for taxonomy determination. Here, we report the complete mitochondrial genome sequence of P. similis, which will provide a better insight into the taxonomic classification and phylogenetic relationship of Phascolosoma.
The tissue samples of P. similis from three individuals was collected from Beibu Bay, China (Beihai, 21.488181 N, 109.532421 E), and the whole body specimens (#GG0152) were deposited at Marine biological Herbarium, Guangxi Institute of Oceanology, Beihai, China. The total genomic DNA was extracted from the muscle of the specimens using an SQ Tissue DNA Kit (OMEGA, Guangzhou, China) following the manufacturer’s protocol. DNA libraries (350 bp insert) were constructed with the TruSeq NanoTM kit (Illumina, San Diego, CA) and were sequenced (2 × 150 bp paired-end) using HiSeq platform at Novogene Company, China. Mitogenome assembly was performed by MITObim (Hahn et al. Citation2013). The complete mitogenome of Phascolosoma esculenta (GenBank accession number: MG873458) was chosen as the initial reference sequence for MITObim assembly. Gene annotation was performed by MITOS (http://mitos2.bioinf.uni-leipzig.de/).
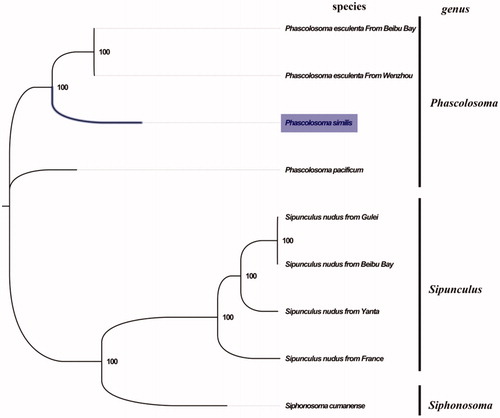
The complete mitogenome of P. similis from Beibu Bay was found to be 16,237 bp in length (GenBank accession number: MN813482), including the usual set of gene for mitogenome except for extra threonine tRNA consisting of 13 protein-coding, 23 tRNA, and 2 rRNA genes, and a putative control region. The overall base composition of the mitogenome was estimated to be A 29.9%, T 33.6%, C 22.6%, and G 13.9%, with a high A + T content of 63.5%, which is slightly lower than Phascolosoma esculenta (65.6%) (Zhao et al. Citation2019). The mitogenomic phylogenetic analyses showed that P. similis was first clustered with P. esculenta and then with P. pacificum in the genus Phascolosoma clade (), which indicated the close relationship between P. similis and P. esculenta. Our result was consistent with the taxonomic classification of Phascolosoma using morphological characteristics (Li Citation1989), which will contribute to further phylogenetic and taxonomic study in the genus Phascolosoma.
Figure 1. Phylogenetic tree in Sipuncula. The complete mitogenomes is downloaded from GenBank and the phylogenic tree is constructed by maximum-likelihood method with 100 bootstrap replicates. The bootstrap values were labeled at each branch nodes. The gene's accession number for tree construction is listed as follows: Phascolosoma pacificum (NC_031412), Phascolosoma esculenta From Wenzhou(NC_012618), Phascolosoma esculenta From Beibu Bay (MG873458), Sipunculus nudus from France (NC_011826), Sipunculus nudus from Yanta (KP751904), Sipunculus nudus from Gulei (KJ754934), Sipunculus nudus from Beibu Bay (MG873457), and Siphonosoma cumanense (MN813483).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cutler E, Cutler N. 1983. An examination of the Phascolosoma subgenera Antillesoma, Rueppellisoma and Satonus (Sipuncula). Zool J Linnean Soc. 77(2):175–187.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Li FL. 1989. Studies on the genus Phascolosoma (Sipuncula) off the China coasts. J Ocean Univ Qingdao. 19:78–90.
- Schulze A, Maiorova A, Timm LE, Rice ME. 2012. Sipunculan larvae and “cosmopolitan” species. Integr Comp Biol. 52(4):497–510.
- Zhao YF, Zhang Q, Zhong SP. 2019. The complete mitochondrial genome of Chinese endemic specie Phascolosoma esculenta (Sipuncula, Phascolosomatidae) from Beibu Bay. Mitochondrial DNA Part B. 4(1):380–381.
