Abstract
Prunus campanulata was a native flowering cherry tree with burgundy red flowers in Taiwan. In this study, the complete chloroplast (cp) genome of P. campanulata was determined. The length of P. campanulata cp genome is 157,938 bp. A total of 130 genes are comprised of 86 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content of cp genome was 36.72%. A maximum-likelihood phylogenetic analysis revealed that P. campanulata is closed to Prunus serrulata. The cp genome sequence is useful for further genetic, evolutionary, and breeding researches.
Keywords:
Prunus campanulata is a species of flowering cherry that belongs to the Rosaceae. P. campanulata is a native species in Taiwan, China, as well as in Vietnam and Japan. In Taiwan, it naturally distributes at the elevation of 300–2000 m and the colors of the flower are more various than other countries. The colors of the flower are from pink (RHS70C, RHS Color Chart) to burgundy red (RHS71A, RHS Color Chart). It infers that P. campanulata has a higher ornamental value and advantage of the breeding potential for low-altitude cultivation (Kato et al. Citation2014). The common name of P. campanulata is Wushe cherry in Yangmingshan national park in Taiwan. The chloroplast genomes are important for phylogenetic and evolutionary researches, but never complete chloroplast sequences are available for P. campanulata on public databases. In this study, we obtained and released the chloroplast genome of P. campanulata by next-generation sequencing and analyzed its phylogenetic status in Prunus.
P. campanulata leaves were collected from Dasyueshan in Taiwan (24°15′57.72′′N, 120°59′55.92′′ E) and a voucher specimen (TGB_PCS11) was deposited in Herbarium of Tree Germplasm Bank, Taiwan Forestry Research Institute. The leaves were used for DNA extraction (Doyle and Doyle Citation1987). The sequencing was conducted at National Yang-Min University, Taiwan and followed the same procedures as our previous reports (Wu et al. Citation2015, Citation2017). The reference-based (Prunus yedoensis (KP732472.1)) assembly was used with CLCbio (Genomics Workbench 8.0.1, CLC Inc., Aarhus, Demark). The annotation was predicted by using GeSeq in default setting (Tillich et al. Citation2017) with manual edition and was submitted to the GenBank (accession number: MH491529).
The complete P. campanulata chloroplast genome was a double-stranded circular DNA of 157,938 bp in length with 244.31× mean coverage. The chloroplast genome contained a pair of inverted repeats (26,436 bp each) separated by a large single-copy region of 85,947 bp and a small single-copy region of 19,119 bp. The overall base composition is 31.16% for A, 32.12% for T, 18.71% for C, 18% for G. The GC content of the whole genome is 36.72%. The genome encoded 130 genes, consisting of 86 coding genes, 8 rRNA genes and 36 tRNA genes. Among all 86 coding genes, 8 genes (rps16, atpF, rpoC1, rpl2, ndhB, ndhA, ndhB, and rpl2) contain one intron, 2 genes (ycf3 and clpP) contain two introns. Seven tRNA genes (trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC, trnA-UGC, and trnI-GAU) contain one intron. Eighteen genes were duplicated in the inverted repeat regions including 7 coding genes (rps19, rpl2, rpl23, ycf2, ndhB, rps7, and ycf1), 7 tRNAs (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU) and 4 rRNAs (rrn16, rrn23, rrn4.5, and rrn5).
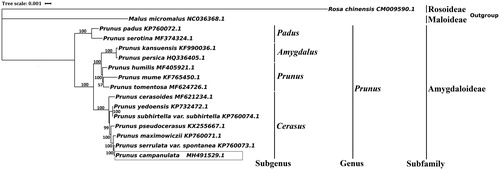
Available chloroplast genomes in Prunus and two Rosaceae species sequences (outgroup) were used for phylogenetic analysis. The MAFFT (Katoh and Standley Citation2013) was used for alignment analysis. The maximum-Likelihood analysis was performed with RAxML (Stamatakis Citation2014) using 1000 bootstrap replicates. The phylogenetic tree was drawn by iTOL (Letunic and Bork Citation2007). The phylogenetic tree () shows that P. campanulata was related to Prunus serrulata var. spontanea and under the clade of Cerasus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Kato S, Matsumoto A, Kensuke Y. 2014. Origins of Japanese flowering cherry (Prunus subgenus Cerasus) cultivars revealed using nuclear SSR markers. Tree Genet Genomes. 10(3):477–487.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23(1):127–128.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wu CC, Chu FH, Ho CK, Sung CH, Chang SH. 2017. Comparative analysis of the complete chloroplast genomic sequence and chemical components of Cinnamomum micranthum and Cinnamomum kanehirae. Holzforschung. 71(3):189–197.
- Wu CC, Ho CK, Chang SH. 2015. The complete chloroplast genome of Cinnamomum kanehirae Hayata (Lauraceae). Mitochondrial DNA Part A. 27(4):2681–2682.