Abstract
Heveochlorella hainangensis is an endophytic green alga in the rubber tree. Its mitocondrion genome was sequenced and characterized. The complete mitogenome contains 54,084 bp with a G + C content of 30.94%. It contains 56 genes, including 32 protein coding genes, 21 tRNA, and 3 rRNA genes. Phylogenetic analysis using the mitogenomes of Trebouxiophyceae species indicated that H. hainangensis is closely related to H. roystonensis, and they both clustered in the Watanabea clade.
Heveochlroella hainangensis J. Zhang is a unicellular Chlorella-like green alga isolated from the rubber tree (Hevea brasiliensis). This alga was found between bark and xylem of the rubber tree young shoots with unknown biological functions (Zhang et al. Citation2008). The type strain FGG01 was isolated from a rubber tree in Chengxi, Haikou, Hainan Province, China (19.983056 N, 110.326944E), and stored at the ClonBank of Institute of Tropical Bioscience and Biotechnology at −80 °C in 15% glycerol. It is also stored at the SAG Culture Collection of Algae (SAG_2360). Due to limited morphological characters for classification of the unicellular microalgae, genome sequences are desired for phylogenetic taxonomy. In this study, we sequenced the complete mitochondrial genome of the type strain of H. hainangensis.
The total genomic DNA was extracted using a genomic DNA extraction kit (Tiangen Biotech, Beijing, China), and was sequenced with both PacBio RSII and Illumina Hiseq 2000 platforms at Genoseq (Wuhan, China). The filtered reads were assembled using CANU (Koren et al. Citation2017) and GATK (Zhu et al. Citation2015). The mitochondrial genome was deposited in the GenBank under accession number MN966687. The completeness of the mitogenome was verified by PCR amplification of the sequence ends. The complete mitochondrial genome contains 54,084 bp with a G + C content 31.0%, which is lower than its close relative H. roystonensis (34.2%). The mitogenome contains 32 protein-coding genes, including 10 ribosomal protein genes, nine NAD(P)H-quinone oxidoreductase (nad) genes, three ATP synthase genes, three cox genes, one cob gene, two intron-encoded endonuclease genes and one tatC gene. It contains 21 tRNA genes, in which two genes (trnM and trnF) are duplicated, whereas the threonine tRNA gene (trnT) was not identified. Three ribosomal RNA genes (rrnL, rrnS, and rrn5) were identified.
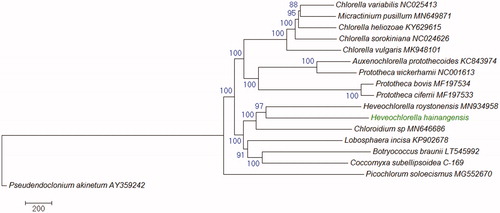
Phylogenetic analysis using the mitochondrial genomes of Trebouxiophyceae species indicated that H. hainangensis is closely related to H. roystonensis, and they both clustered together with Chloroidium sp. in the Watanabea clade (), which is in agreement with previous reports (Ma et al. Citation2013; Zhang et al. Citation2008).
Figure 1. Phylogenetic relationships of Trebouxiophyceae species based on their mitogenomes. The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Evolutionary analyses were conducted in MEGA7 (Kumar et al. Citation2016). The tree is drawn to scale as indicated by the scale bar and rooted with a Ulvophyceae species Pseudendoclonium akinetum as an outgroup. GenBank accession numbers are shown behind the taxon names.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Ma S, Huss VAR, Tan D, Sun X, Chen J, Xie Y, Zhang J. 2013. A novel species in the genus Heveochlorella (Trebouxiophyceae, Chlorophyta) witnesses the evolution from an epiphytic into an endophytic lifestyle in tree-dwelling green algae. Eur J Phycol. 48(2):200–209.
- Zhang J, Huss VAR, Sun X, Chang K, Pang D. 2008. Morphology and phylogenetic position of a trebouxiophycean green alga (Chlorophyta) growing on the rubber tree, Hevea brasiliensis, with the description of a new genus and species. Eur J Phycol. 43(2):185–193.
- Zhu P, He L, Li Y, Huang W, Xi F, Lin L, Zhi Q, Zhang W, Tang YT, Gen C, et al. 2015. Correction: OTG-snpcaller: an optimized pipeline based on TMAP and GATK for SNP calling from ion torrent data. PLoS One. 10(9):e0138824.
