Abstract
Sinomenium acutum is a woody vine of great virtue in traditional Chinese medicine and, nowadays, a potent anticancer drug against several types of cancers. In the present study, we have assembled and characterized the very first chloroplast (cp) genome sequence of S. acutum. The whole chloroplast genome is 162,966 bp in length, containing a pair of inverted repeats (IR) of 25,051 bp each, separated by one large single-copy region (LSC, 91,627 bp) and a small single-copy region (SSC, 21,237 bp). The cp genome encodes 132 unique genes that contain 85 protein-coding genes, 37 transfer RNA genes, and 8 ribosomal RNA genes. In our phylogeny of Ranunculales, Menispermum dauricum is sister to S. acutum.
Sinomenium acutum is a deciduous climber with woody twining stems native to East Asia. It was first recorded as an herbal medicine in Ben-Cao-Gang-Mu (Compendium of Materia Medica). Known as Qing-Feng-Teng in traditional Chinese medicine (TCM), it has been used for hundreds of years for the treatment of rheumatism and neuralgia (Zhao et al. Citation2012). Sinomenium acutum contains the alkaloid sinomenine, an isoquinoline that can be used to treat rheumatoid arthritis and other inflammatory conditions (Xu et al. Citation2008). Sinomenine exhibits neuroprotection in brain injury (Nishida and Satoh Citation2006). Moreover, sinomine has potential anti-cancer activities against breast cancer, colon cancer, lung cancer, liver cancer, and gastric cancer (Yang et al. Citation2018). Owing to its immense virtue in medicine, we have characterized the very first complete chloroplast genome sequence of S. acutum.
Fresh leaf materials of S. acutum were collected from China, Anhui Province, Xiuning County, Qiyunshan. The voucher specimen (Pan Li LP196752) was deposited in the Zhejiang University Herbarium (HZU). Silica gel dried leaves were used to extract DNA, using DNA Plantzol Reagent (Invitrogen, Shanghai, China) following the manufacturer’s protocol. NOVOPlasty (Dierckxsens et al. Citation2016) was used to construct the complete chloroplast genome through de novo assembly and map to reference genome Menispermum dauricum (MH298220; Hina et al. Citation2018). The whole sequence was annotated with Geneious v 11.0.2 (Biomatters Ltd., Auckland, New Zealand) and then submitted to GenBank MT040976.
The whole chloroplast genome of S. acutum is 162,966 bp in length, with one large single-copy region (LSC, 91,627bp), a small single-copy region (SSC, 21,237bp), and two inverted repeats (IR: 25,051bp each). The overall GC content of the genome is 38%. The cp genome consists of a total of 132 genes with 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. There are 19 genes (8 tRNA genes and 11 protein-coding genes) with one intron and 3 genes (rps12, clpP, and ycf3) with two introns. Ycf1 and rps19 are the pseudogenes crossing the SSC/IR and LSC/IR borders. The rps12 gene is trans-spliced, with the 5′ end located in the LSC and the 3′ end duplicated in the IR regions.
The phylogenetic tree was constructed using both maximum-likelihood (ML) and Bayesian Inference (BI) methods, with Sabia yunnanensis (KU204902) as an outgroup. ML analysis was carried out on XSEDE v8.2.10 using RAxML-HPC2 (Stamatakis Citation2014). BI analysis was run with MrBayes on XSEDE v3.2.6 (Ronquist and Huelsenbeck Citation2003). Both ML and BI analyses were implemented on the CIPRES Science Gateway V. 3.3 (Miller et al. Citation2012) and generated the same tree topology (). Within Menispermaceae, S. acutum is sister to M. dauricum.
Figure 1. Molecular phylogeny of Ranunculales based on 23 complete cp genomes. The accession numbers are listed as below: Aconitum angustius (MF155664); Akebia quinata (KX611091); Anemoclema glaucifolium (MG010811); Berberi samurensis (KM057374); Circaeaster agrestis (KY908400); Clematis terniflora (KJ956785); Coptis chinensis (KY120323); Coreanomecon hylomeconoides (KT274030); Decaisnea insignis (KY200671); Epimedium lishihchenii (KU522472); Euptelea pleiosperma (KU204900); Gymnaconitum gymnandrum (KT964697); Kingdoniauni flora (KY908401); Megaleranthis saniculifolia (FJ597983); Menispermum dauricum (MH298220); Nandina domestica (DQ923117); Papaver somniferum (KU204905); Ranunculus occidentalis (KX557270); Sinomenium acutum (MT040976); Sinopodophyllum hexandrum (KR779994); Stephania japonica (KU204903); Trollius chinensis (KX752098); Vancouveria planipetala (MH337373); Sabia yunnanensis (KU204902).
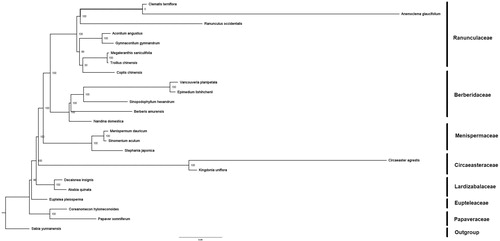
In conclusion, this is the first report of the complete cp genome of medicinally important liana S. acutum, and will be valuable for future phylogenomic studies.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hina F, Jin Z, Yang Z, Li P, Fu C. 2018. The complete chloroplast genome of Menispermum dauricum (Menispermaceae, Ranunculales). Mitochondrial DNA Part B. 3(2):913–914.
- Miller MA, Pfeiffer W, Schwartz T. 2012. The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the campus and beyond. July 16–19; New York (NY): ACM. p. 39.
- Nishida S, Satoh H. 2006. In vitro pharmacological actions of sinomenine on the smooth muscle and the endothelial cell activity in rat aorta. Life Sci. 79(12):1203–1206.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xu M, Liu L, Qi C, Deng B, Cai X. 2008. Sinomenine versus NSAIDs for the treatment of rheumatoid arthritis: a systematic review and meta-analysis. Planta Med. 74(12):1423–1429.
- Yang S, Peng L-Y, Peng W, Huang C, Wei D-N, Mou M-T, Liang F, Gao, Y-X. 2018. Anticancer potentials of sinomenine from Sinomenium acutum: A mini-review. Trop J Pharm Res. 17(12):2519–2526.
- Zhao X-X, Peng C, Zhang H, Qin L-P. 2012. Sinomenium acutum: a review of chemistry, pharmacology, pharmacokinetics, and clinical use. Pharm Biol. 50(8):1053–1061.
