Abstract
Rubus chingii Hu is an important traditional Chinese medicine with a beneficial effect on the kidney. In this study, we report the complete chloroplast genome sequence of R. chingii. The assembled chloroplast genome was 155843 bp in length, containing two inverted repeated (IR) regions of 25372 bp each, a large single copy (LSC) region of 86456 bp and a small single copy (SSC) region of 18643 bp. The genome contained 130 genes, including 86 protein genes, eight rRNA genes, and 36 tRNA genes. The overall GC content of complete chloroplast genome was 37.1%. Phylogenetic analysis demonstrated that R. chingii clustered together with the monophyletic group of R. crataegifolius and R. takesimensis, suggesting a close relationship among the three species from Tribe Rubeae.
The Chinese wild raspberry Rubus chingii Hu is a widely distributed economic and medicinal plant belonging to the Rubus genus in the family Rosaceae (Gu et al. Citation1993). The fruits of R. chingii have been employed as a functional food as well as a traditional medicine to treat kidney enuresis and other illnesses in the clinic (He et al. Citation2018). The fresh leaves of this species were often used as popular beverages in China. The ethanol extract from the fruits of R. chingii exhibited protecting activity to the skin against UVB damage, suggesting potential applications in cosmetics (She et al. Citation2019). Furthermore, Rubus is one large and diverse genus containing plentiful species with similar morphologies, causing complicated arguments on the species identification and classification (Alice and Campbell Citation1999). Molecular identification is an efficient approach for species-level differentiation via standard DNA barcodes. Here, we report the complete chloroplast genome of R. chingii to provide genomic resources for molecular marker development and clarify the phylogenetic position of this plant within others in Rosaceae.
The sample of Rubus chingii was collected from the area of Jiulongshan National Nature Reserve in Zhejiang Province (28°41′58.49″N, 119°10′51.01″E). The specimen was deposited in the collection center of Zhejiang Chinese Medical University with the specific identifying number of JSL-1903. Total genomic DNA was extracted and sequenced using the Illumina Hiseq Platform according to the previous reprot (Ying et al. Citation2019). The chloroplast genome of R. chingii was assembled by metaSPAdes with the chloroplast sequence of R. crataegifolius as reference (Nurk et al. Citation2017). The chloroplast was annotated using GeSqe and further corrected by BLAST (Tillich et al. Citation2017). The complete cp genome of R. chingii was submitted to GenBank with the accession number of MN885523.
The length of the complete chloroplast genome sequence of R. chingii was 155843 bp, with a large single copy (LSC) region of 86456 bp, a small single copy (SSC) region of 18643 bp, and two separated inverted repeated (IR) regions of 25372 bp each. A total of 130 genes were identified in the cp of R. chingii, including 86 protein-coding genes, 36 tRNA genes and 8 rRNA genes. The overall GC content was 37.1%, and the corresponding contents for LSC, SSC and IR regions were 35.0%, 30.9% and 42.8%, respectively. The genome included 16 duplicated genes in the IR region and exhibited 51.5% protein-coding sequences. Moreover, a total of 57 small single repeats (SSR) are identified in the cp of R. chingii, ranging from 10 bp to 18 bp.
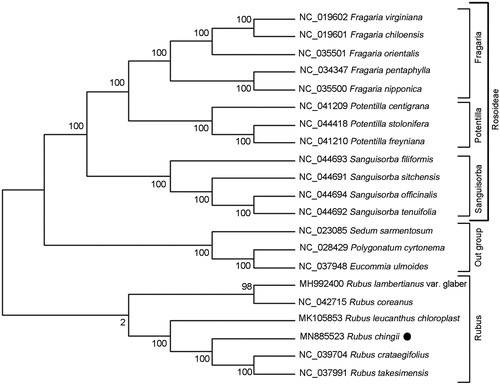
The complete genome sequences of R. chingii and other 17 representative species from Rosaceae family were analyzed using MEGA 7.0 by maximum-likelihood (ML) method to confirm the phylogenetic position (Kumar et al. Citation2016). The result demonstrated a sister relationship between R. chingii and the combined group forming by R. crataegifolius and R. takesimensis, suggesting a close relationship among the three species (). In addition, Rubus species clustered in a clade together with high supporting scores, but did not display sister-relationship with the monophyletic group of other species in Rosaceae (). These results suggested the unique evolutionary status of Rubus and required further revision on the taxonomy of Family Rosaceae.
Figure 1. ML phylogenetic tree analysis of Rubus chingii and other representative Rosaceae plants based on the complete chloroplast genome sequences. Numbers on the nodes are bootstrap values from 100 replicates. The GenBank accession numbers were listed before the species name. The species of Sedum sarmentosum, Eucommia ulmoides and Polygonatum cyrtonema were used as the outgroup species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alice LA, Campbell CS. 1999. Phylogeny of Rubus (Rosaceae) based on nuclear ribosomal DNA internal transcribed spacer region sequences. Am J Bot. 86(1):81–97.
- Gu Y, Zhao CM, Jin W, Li WL. 1993. Evaluation of Rubus germplasm resources in China. Acta Hortic. 352:317–324.
- He Y, Jin S, Ma Z, Zhao J, Yang Q, Zhang Q, Zhao Y, Yao B. 2018. The antioxidant compounds isolated from the fruits of Chinese wild raspberry Rubus Chingii Hu. Nat Prod Res. 1–4. doi:10.1080/14786419.2018.1504046
- Kumar S, Stecher G, Tamura K. 2016. Mega 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner P A. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- She JM, Yuan HH, Zhang J, Si ZN, Lan MB. 2019. Protective effect of the Rubus chingii Hu. fruit extract on ultraviolet B-induced photoaging via suppression of mitogen-activated protein kinases (MAPKs) in vitro. Arch Biol Sci (Beogr)). 71(3):541–550.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Ying Z, Wang Q, Yu S, Liao G, Ge Y, Cheng R. 2019. The complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Lysimachia hemsleyana. Mitochondrial DNA B. 4(2):3878–3879.
