Abstract
The length of the complete mitochondrial genome was 16,626 bp for Ophiura kinbergi, containing 35 genes similar to other known ophiuroids. The GC content was 33.35%. The gene structure was identical to known mitochondrial genomes from the species of genus Ophiura. Phylogenetic results indicated conserved relationships between O. kinbergi and three other Ophiura species. These results could provide detailed information for better understanding the evolution patterns of brittle stars, especially for the genus Ophiura.
Keywords:
Ophiura kinbergi, belonging to Ophiurida, Ophiuroidea, is a common species widely distributed from the Red Sea to the West Pacific Ocean (Liao Citation2004). Marine bioactive substances could be produced by this species, such as Ophiurasaponin and some anti-oxidants (Zhen et al. Citation2015). The sample was collected from the Yellow Sea (35°N, 123°E) and was stored in Key Laboratory of Science and Engineering for Marine Ecology and Environment, First Institute of Oceanography, MNR (No. FIO-ECH-YSJ01).
The whole genome of O. kinbergi was sequenced on the Illumina HiSeq 2500 Sequencing Platform (Illumina, Hayward, CA) by Novogene Corporation (Beijing, China). It was assembled using SPAdes 3.6.1(Bankevich et al. Citation2012). BLAST algorithm-based search against other available Ophiuroidea mitogenomes in GenBank was used to query mitochondrial genomic fragments. Price (Ruby et al. Citation2013) and MITObim v1.8 (Hahn et al. Citation2013) were used to fill the gaps. The mitochondrial reads were collected by bowtie2 (Langmead and Salzberg Citation2012) and reassembled. The circular structure of the mitochondrial genome was verified using Bandage (Wick et al. Citation2015). The complete mitochondrial genome of O. kinbergi was submitted to Genbank and the accession number was MH910618.
The gene structure of O. kinbergi contains 35 genes, including 13 coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. Lengths of the whole mitochondrial genomes and protein-coding genes were 16,626 and 11,535 bp, respectively. The GC content of this mitochondrial genome was 33.35%. The gene order within the genus Ophiura was identical, which illustrated the conserved gene structure in this genus.
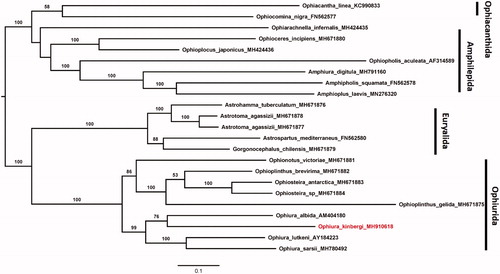
Phylogenetic relationships among O. kinbergi and 22 brittle stars were analyzed using the maximum-likelihood method by IQTREE (Nguyen et al., 2015). The nucleotide models of each gene were selected by ModelFinder (Kalyaanamoorthy et al. Citation2017). This phylogenetic tree was divided into four orders, which were consistent with O’Hara et al. (Citation2018) (). The relationship between O. kinbergi and other ophiuroids showed it closest to Ophiura albida. This study provided additional details for the phylogenetic studies of Ophiuroidea and genus Ophiura.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Meth. 14(6):587–589.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Meth. 9(4):357–359.
- Liao YL. 2004. Fauna Sinica: Invertetrata Vol. 40, Echinodermata Ophiuroidea. Beijing, China: Science Press.
- O’Hara TD, Stöhr S, Hugall AF, Thuy B, Martynov A. 2018. Morphological diagnoses of higher taxa in Ophiuroidea (Echinodermata) in support of a new classification. EJT. 416:1–35.
- Ruby GJ, Bellare P, Derisi JL. 2013. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. Genes, Genomes, Genetics. 3:865–880.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Zhen J, Cong J, Qu J, Hou J, G. Ch 2015. Research on antioxidant and antibacterial activity of polysaccharide in Ophiura kinbergi. China Brewing. 34(11):136–139. (in Chinese with English abstract)