Abstract
We determined the complete mitochondrial DNA sequence of Silurus lithophilus, a catfish endemic to Lake Biwa, Japan. The mitogenome was 16,524 bp in length and the gene contents and order agreed with those of typical vertebrates. A molecular phylogenetic analysis based on 12 published Silurus mitogenomes showed that the mitogenome of S. lithophilus (LC520058) was closest to that of Silurus asotus from Japan (AP012022). These two mitogenomes were identical in the short (ca. 170 bp) gene region widely used in metabarcoding for environmental DNA analysis of fishes.
Silurus lithophilus is a catfish species endemic to Lake Biwa, central Japan, inhabiting rocky and gravel slopes of the lake (Tomoda Citation1962). It is consumed locally, but it is classed as a near-threatened species in the Japanese Red Data Book (Ministry of the Environment Citation2019). Although a molecular phylogenetic study, including partial mitochondrial DNA sequences, of this species has already been published (Tabata et al. Citation2016), its complete mitochondrial genome (mitogenome) has not been reported.
In this study, we determined the mitogenome sequence of a specimen of S. lithophilus collected north of Port Onoe, Shiozu Bay, in Lake Biwa. The fish was purchased from a local fisherman, and the body specimen was deposited in the Kanagawa Prefectural Museum of Natural History, Japan, under registration number KPM-NI 45274. Genomic DNA was isolated from a fin clip and sequenced using an Illumina MiSeq Sequencer. The resulting reads were assembled using CLC Genomics Workbench (ver. 11.01; Qiagen, Hilden, Germany). The genes were annotated with MitoAnnotator (Iwasaki et al. Citation2013).
The mitogenome sequence obtained (DDBJ accession no. LC520058) was 16,524 bp in length and included 22 transfer RNAs, 2 ribosomal RNAs (rRNAs), 13 protein-coding genes, and a D-loop region, with the order identical to that of typical vertebrates. Using the entire sequence (15,635 bp) except the D-loop, the phylogenetic relationships among S. lithophilus and 12 available Silurus mitogenomes were analyzed using the maximum-likelihood (ML) method (for details, see the figure legend). In the resulting tree (), the mitogenome of S. lithophilus was phylogenetically closest to that of Silurus asotus from Japan (AP012022).
Figure 1. Maximum-likelihood (ML) tree showing the phylogenetic position of the mitogenome of Silurus lithophilus (LC520058) among 12 available Silurus mitogenomes, with two Kryptopterus ones as outgroups. The dataset includes the entire mitogenome except the D-loop area, which showed alignment ambiguities; the resulting dataset was 15,655 bp in length. The ML analysis was performed using MEGA 7 (Kumar et al. Citation2016) with the General Time Reversible Substitution Model, rates among sites Gamma distributed with Invariant sites, and 500 bootstrap replicates. Bootstrap support values are indicated at the nodes of the tree. Countries where the specimens were collected are on the right side of the figure.
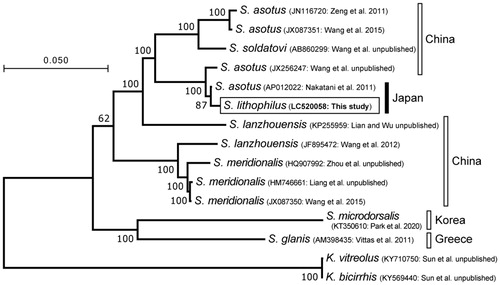
The nucleotide identity between the two closely related mitogenomes was 99.49% (16,440 of 16,525 bp). Although one base substitution (including an insertion/deletion) was observed every 194 bases on average, there was no substitution in the ca. 170 bp ‘MiFish’ region in the 12S rRNA gene, which is widely used as a metabarcoding region for environmental DNA analysis of fishes (Miya et al. Citation2015). To distinguish the two Japanese catfishes using similar-length DNA fragments, it is necessary to analyze more variable regions. The following eight 180-bp regions are good candidates because of their high variabilities (contain more than four substitutions): 2701–2880 (four substitutions), 2761–2940 (four substitutions), 4381–4560 (five substitutions), 6121–6300 (four substitutions), 15,721–15,900 (six substitutions), 15,781–15,960 (seven substitutions), 15,841–16,020 (seven substitutions), and 15,901–16,080 (five substitutions) (nucleotide positions based on the aligned 16,525 bp). Of the eight regions, the last four overlap sequentially and overall within the D-loop region, while the first two are within the region from the end of the 16S rRNA gene to the beginning of ND1. The remaining two regions, 4381–4560 and 6121–6300, are within ND2 and COI, respectively.
Geolocation information
35°28′25.1′′N; 136°10′23.5′′E
Acknowledgments
We thank Hiroshi Senou (Kanagawa Prefectural Museum for Natural History) for registering our specimen in the KPM fish collection. Our special thanks go to Masatomi Matsuoka for help obtaining the specimen.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Ministry of the Environment. 2019. The Japanese Red List of Threatened Species, 2019 version. [accessed 2020 Feb]. https://www.env.go.jp/press/files/jp/110615.pdf.
- Miya M, Sato Y, Fukunaga T, Sado T, Poulsen JY, Sato K, Minamoto T, Yamamoto S, Yamanaka H, Araki H, et al. 2015. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R Soc Open Sci. 2(7):150088.
- Nakatani M, Miya M, Mabuchi K, Saitoh K, Nishida M. 2011. Evolutionary history of Otophysi (Teleostei), a major clade of the modern freshwater fishes: Pangaean origin and Mesozoic radiation. BMC Evol Biol. 11(1):e177.
- Park CE, Park Y-J, Kim M-C, Park M-K, Jung YG, Choi S-D, Jo YJ, Kang G-U, Kim M-J, Li QX, et al. 2020. The first complete mitochondrial genome sequence of the korean endemic catfish Silurus microdorsalis (Actinopteri, Siluriformes, Siluridae). Mitochondr DNA B. 5(1):131–132.
- Tabata R, Kakioka R, Tominaga K, Komiya T, Watanabe K. 2016. Phylogeny and historical demography of endemic fishes in Lake Biwa: the ancient lake as a promoter of evolution and diversification of freshwater fishes in western Japan. Ecol Evol. 6(8):2601–2623.
- Tomoda Y. 1962. Studies of the fishes of Lake Biwa-ko―I Morphological study of the three species of catfishes of the genus Parasilurus from Lake Biwa-ko, with reference to their life. Jap J Ichthyol. 8:126–146.
- Vittas S, Drosopoulou E, Kappas I, Pantzartzi CN, Scouras ZG. 2011. The mitochondrial genome of the European catfish Silurus glanis (Siluriformes, Siluridae). J Biol Res (Thessalon). 15:25–35.
- Wang QR, Xu C, Xu CR, Wang RJ. 2012. Mitochondrial genome structure of yellow river catfish (Silurus lanzhouensis) and phylogenetic analysis. Acta Sci Nat Univ Pekinensis. 48:376–380.
- Wang QR, Xu C, Xu CR, Wang RJ. 2015. Complete mitochondrial genome of the Southern catfish (Silurus meridionalis Chen) and Chinese catfish (S. asotus Linnaeus): structure, phylogeny, and intraspecific variation. Genet Mol Res. 14(4):18198–18209.
- Zeng Q, Wang ZJ, Peng ZG. 2011. Mitochondrial genome of Silurus asotus (Teleostei: Siluriformes). Mitochondr DNA. 22(5–6):162–164.
