Abstract
The whole mitochondrial genome sequence of Lepturichthys fimbriata was amplified by primer walking sequence method, and then the whole genome genetic map of mitochondrion was obtained and mapped. The mitochondrial genome of L. fimbriata is a double-strand ring-like molecule with a total length of 16,567bp. It contains 2 rRNA genes (12SrRNA and 16SrRNA), 13 protein-coding genes (COI, COII, COIII, ATP6, ATP8, ND1, ND2, ND3, ND4, ND4L, ND5, ND6, Cytb), 22 tRNA coding genes, and one non-coding control region (D-loop control region). The overall nucleotide composition was A: 30.4%, T: 24.4%, C: 28.7%, and G: 16.5%, exhibiting an A + T bias of 54.8%. Phylogeny showed the phylogenetic relationships of different genera in the family Balitoridae and found the genus Lepturichthys is clustered with the genus Jinshaia, which suggested the intergeneric hybridization has likely occurred in both lineages.
Lepturichthys fimbriata is endemic to China and mainly distributed in jinsha river, belonging to the genus Lepturichthy within the family Balitoridae (Ding Citation1994). It is a kind of benthic small fish inhabiting in the rocky beach of the mountain stream and river, and living in the rocky beach of the river and rapids. (Ding Citation1994). Until now, there are seventeen mitochondrial genomes of family Balitoridae available in NCBI database. However, the phylogenetic relationships of family Balitoridae have been not studied. In this study, the mitochondrial genome of L. fimbriata was determined, and the phylogenetic tree was constructed to explore the phylogenetic relationships of family Balitoridae.
Lepturichthys fimbriata in the experiment was collected in Yibin section of upper reaches of the Yangtze River (104°58′E, 28°72′N), Sichuan Province in 2012. After morphological identification, the specimen was deposited in our laboratory (Specimen number:201211281003, Animal genetics lab, Jianghan University). The tail fin was cut off and stored in 95% ethanol. In order to amplify the complete mitochondrial genome of L. fimbriata, seventeen sets of primers were designed using Primer5 from the conservative regions according to the alignments of seventeen complete mitochondrial genomes available within the family Balitoridae. Total genomic DNA was extracted by standard phenol-chloroform methods (Sambrook and Russell Citation2001). SeqMan program in DNAstar software was used to check and assemble mitogenome.
The mtDNA sequence of L. fimbriata is 16,567bp in length (GenBank accession number:MN882155). MitoAnnotator was used to annotate the protein-coding genes, rRNA genes, tRNA genes, and a control region (CR) of mitochondrial genome of L. fimbriata (Iwasaki et al. Citation2013). Gene arrangement and organization of the mitochondrial genome of L. fimbriata are similar to those of other teleosts (Broughton et al. Citation2001; Xu et al. Citation2016). The genome contains two rRNA genes (12 SrRNA and 16 SrRNA) and 13 protein-coding genes (COI, COII, COIII, ATP6, ATP8, ND1, ND2, ND3, ND4, ND4L, ND5, ND6, Cytb), 22 tRNA genes and a non coding control area (D-loop). The overall nucleotide composition was A: 30.4%, T: 24.4%, C: 28.7%, and G: 16.5%, exhibiting an A + T bias of 54.8%. There are six variable positions in the mitochondrial genome of L. fimbriata determined in our lab and the previously reported L. fimbriata mitochondrial genome occurring in the 16 SrRNA, ND4L, ND5 and ND6 genes (Xu et al. Citation2016).
In order to clarify the phylogenetic relationship of the family Balitoridae, the phylogenetic tree based on 13 protein-coding genes (11,349bp) was constructed using Bayesian inference method (Ronquist et al. Citation2012). Phylogeny showed the phylogenetic relationships at genera level in the family Balitoridae (). The genera Metahomaloptera and Homalosoma were located in the basal position followed by Neohomaloptera and Homalopteroides. And then, the clade of (Homaloptera, (Sinogastromyzon, (Lepturichthys, Jinshaia))) was diverged. The genus Lepturichthys is clustered with the Jinshaia species as a sister relationship which suggest the genus Lepturichthys is closely related to the genus Jinshaia. In addition, the homology of L. fimbriata and Jinshaia sinensis was as high as 98.15% via BLAST comparison in NCBI and indicated that intergeneric hybridization has likely occurred between Lepturichthys and Jinshaia lineages. This result confirmed the conclusion of Tang et al. (Citation2012).
Figure 1. Phylogenetic tree of balitorine loaches reconstructed from 18 mitogenomes of Balitoridae using Bayesian inference method.The sequenced species in this study are shown in red.
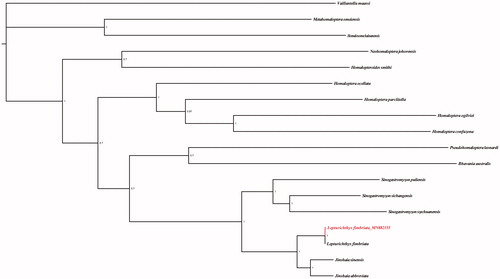
In conclusion, this is the first time to explore the phylogenetic relationship of family Balitoridae from the mitogenomic perspective. The results would facilitate further investigations of molecular systematics and evolution of species in the family Balitoridae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Broughton RE, Milam JE, Roe BA. 2001. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11(11):1958–1967.
- Ding R. 1994. The fishes of Sichuan. Chengdu, Sichuan, China. Sichuan: Publishing House of Science and Technology (in Chinese).
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh T. P, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Ronquist F, Teslenko M, van der Mark P, Ayres D L, Darling A, Höhna S, Larget B, Liu L, Suchard M A, Huelsenbeck J P, et al. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. New York: Cold Spring Harbor Laboratory Press.
- Tang QY, Liu SQ, Yu D, Liu HZ, Danley PD. 2012. Mitochondrial capture and incomplete lineage sorting in the diversification of balitorine loaches (Cypriniformes, Balitoridae) revealed by mitochondrial and nuclear genes. Zoologica Scripta. 41(3):233–247.
- Xu DM, Yang Z, Xu N, Xiong MH, Lei P, Que YF. 2016. The complete mitochondrial genome of Lepturichthys fimbriata (Teleostei, Balitoridae, Balitorinae). Mitochondrial DNA A DNA Mapp Seq Anal.. 27(2):1064–1065.
