Abstract
Dendrobium wattii is a rare and ornamental orchid that is placed in Sect. Formosae of Dendrobium. The complete chloroplast (cp) genome sequence and the genome features of D. wattii were reported for new data on the phylogeny of Dendrobium. The complete cp genome sequence of D. wattii is 159,366 bp in length, including a large single-copy region (LSC, 87,192 bp), a small single-copy region (SSC, 18,422 bp), and two inverted repeat sequences (IRs, 26,876 bp, each). The total content of GC is 37.2%. The cp genome contains 133 genes, consisting of 131 unique genes (79 protein-coding genes, 30 tRNAs, and 4 rRNAs). The phylogenetic analysis suggested that D. wattii be closely related to other species of Dendrobium.
As the second-largest genus in Orchidaceae, Dendrobium has been problematic with a focus on generic and infrageneric relationships (Adams Citation2011; Xiang et al. Citation2013). Dendrobium wattii belongs to the Sect. Formosae of Dendrobium, which has a characteristic black hirsute in the leaves and stems and has white showy flowers (Chen et al. Citation2009). In this paper, the complete chloroplast genome of D. wattii was reported in order to provide molecular data for the systematic problems in Dendrobium.
Leaf samples of D. wattii were collected from the Wild Orchid Conservation Center of Yunnan Fengchunfang Biotechnology Co., Ltd., which is located in Fumin County, Yunnan Province, China (25°20′19′′N, 102°27′26′′E). The voucher specimen was deposited in Herbarium of Southwest Forestry University (HSFU, Lilu20180002). The total genomic DNA was extracted from the fresh leaf by using the modified CTAB procedure of Doyle and Doyle (Citation1987) and sequenced on Illumina Hiseq 2500 platform (Illumina, San Diego, CA). Genome sequences were screened out and assembled with MITObim V1.8 (Hahn et al. Citation2013).
The cp genome sequence of D. wattii (GenBank accession MN913340) is 159,366 bp in length, of which the GC content is 37.2%. The sequence is composed of a large single-copy region (LSC, 87,192 bp), a small single-copy region (SSC,18,422 bp), and two inverted repeat sequences (IRs, 26,876 bp, each), while the corresponding values of the GC contents are 35, 30.4, and 43.2%, respectively. The cp genome encodes 133 genes, consisting of 131 unique genes (79 protein-coding genes, 30 tRNAs, and 4 rRNAs). The gene order and organization of the D. wattii are consistent with those of other species in Dendrobium (Yan et al. Citation2016; Huang et al. Citation2019; Pan et al. Citation2019).
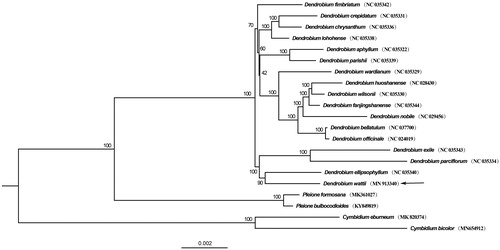
To confirm the systematic position of D. wattii, a phylogenetic tree was constructed based on 72 protein-coding genes from 17 species of Dendrobium, with 4 species from Cymbidium and Pleione as outgroup using maximum-likelihood (ML) method. It was shown that D. wattii was clustered with other species of Dendrobium (). This newly reported cp genome of D. wattii will be helpful for further investigation of taxonomy and phylogeny in Dendrobium.
Figure 1. Phylogenetic position of Dendrobium wattii inferred by maximum-likelihood (ML) based on 72 protein-coding genes from17 species of Dendrobium, with 4 species from Pleione and Cymbidum as outgroup. Other sequences used in this study were downloaded from the NCBI GenBank database. The bootstrap values are shown next to the nodes, with an arrow indicating D. wattii.
Acknowledgments
We are grateful to Dr. Fei Zhao in Kunming Institute of Botany, Chinese Academy of Sciences, for his help in this manuscript. We also thank Mr. Zhi-feng Xu and Mrs. Xiao-yun Wang in the Orchid Conservation Center of Yunnan Fengchunfang Biotechnology Co., Ltd, for providing sample materials.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Adams PB. 2011. Systematics of Dendrobiinae (Orchidaceae), with special reference to Australian taxa. Bot J Linn Soc. 166(2):105–126.
- Chen SC, Liu ZJ, Zhu GH, Lang KY, Ji ZH, Luo YB, Jin XH, Cribb PJ, Wood JJ, Gale SW. 2009. Orchidaceae. In: Wu ZY, Raven PH, Hong D, editors. Flora of China, vol. 25. Beijing (China): Science Press; St. Louis (MO): Missouri Botanical Garden Press, p. 211–235.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Huang ZC, Pan YY, Chen GZ, Chen LJ, Wu XY, Huang J. 2019. The complete chloroplast genome of Dendrobium harveyanum (Orchidaceae). Mitochondrial DNA Part B: Res. 4(2):3200–3201.
- Pan YY, Li TZ, Chen JB, Huang J, Rao WH. 2019. Complete chloroplast genome of Dendrobium thyrsiflorum (Orchidaceae). Mitochondrial DNA Part B: Res. 4(2):3192–3193.
- Xiang XG, André S, Li DZ, Huang WC, Chung SW, Jian W, Li JW, Zhou HL, Jin WT, Lai YJ, et al. 2013. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid andnuclear sequences. Mol Phylogenet Evol. 69(3):950–960.
- Yan W, Niu Z, Zhu S, Ye M, Ding X. 2016. The complete chloroplast genome sequence of Dendrobium nobile. Mitochondrial DNA Part A: DNA Mapp, Seq, Anal. 27(6):4090–4092.
