Abstract
The first complete chloroplast genome of Curcuma zedoaria (Zingiberaceae) was reported in this study. The C. zedoaria chloroplast genome was 162,135 bp in length, and consisted of one large single-copy (LSC) region of 86,966 bp, one small single-copy (SSC) region of 15,737 bp, and a pair of inverted repeat (IR) regions 29,716 bp. It encoded 141 genes, including 87 protein-coding genes (79 PCG species), 46 tRNA genes (28 tRNA species), and 8 rRNA genes (4 rRNA species). The phylogenetic analysis based on single nucleotide polymorphisms strongly supported that C. zedoaria, Curcuma roscoeana, Curcuma longa, and Stahlianthus involucratus clustered together in group Curcuma II within the family Zingiberaceae.
Curcuma zedoaria, commonly known as ‘zedoary’ and ‘white turmeric’ in English and ‘ezhu’ in Chinese, is an important Curcuma with an ancient medicinal history in China and Japan, where the plant is used as a carminative and stomachic (Branney Citation2005; Dosoky and Setzer Citation2018). The essential oil of zedoary essential oil is used in the perfume and liquor industries, and the bitter-tasting rhizome is used in the preparation of some curry powders and eaten as a vegetable in Thailand (Branney Citation2005). Numerous studies have demonstrated that the rhizome of C. zedoaria has many pharmacological effects, such as reproductive toxicity, anti-cancer, anti-inflammatory, anti-allergic, antimicrobial activities, and so on (Matsuda et al. Citation2004; Wilson et al. Citation2005; Makabe et al. Citation2006; Zhou et al. Citation2013). Furthermore, it is also widely cultivated as ornamental plant due to its attractive flowers (Branney Citation2005). However, to the best of our knowledge, the chloroplast genome of C. zedoaria is still unclear.
Curcuma zedoaria was stored at the resource garden of environmental horticulture research institute (23°23′N,113°26′E; specimen accession number Cz 2015001), Guangdong Academy of Agricultural Sciences, Guangzhou, China. Total chloroplast DNA was extracted from C. zedoaria fresh leaves using the modified sucrose gradient centrifugation method (Li et al. Citation2012). Chloroplast DNA (accession number CzDNA2017) was stored at −80 °C in Guangdong key lab of ornamental plant germplasm innovation and utilization, environmental horticulture research institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China. Two libraries were constructed and sequenced on an Illumina Hiseq X Ten instrument and a PacBio Sequel platform (Biozeron, Shanghai, China), respectively. The Illumina and PacBio sequencing data were deposited in the NCBI sequence read archive under accession numbers SRR8189801 and SRR8184503, respectively. After trimming, the Illumina sequencing and PacBio sequencing yielded 57.1 M clean data of 150 bp paired-end reads and 0.54 M clean data of 8–10 kb subreads, respectively. The chloroplast genome of C. zedoaria was assembled and annotated by using the reported method (Li et al. Citation2019). The complete chloroplast genome sequence of C. zedoaria was submitted to GenBank (accession number: MK262734).
The complete chloroplast genome of C. zedoaria was 162,135 bp in length, and comprised a pair of inverted repeat (IR) regions of 29,716 bp each, a large single-copy (LSC) region of 86,966 bp, and a small single-copy (SSC) region of 15,737 bp. It was predicted to contain a total of 141 genes, including 46 tRNAs (28 tRNAs species), 87 protein-coding genes (79 PCG species) and 8 rRNAs (4 rRNAs species). Twenty gene species occurred in double copies and were located in the IR regions, including 8 PCG genes (ndhB, rpl2, rpl23, rps7, rps12, rps19, ycf1, ycf2), 8 tRNAs (trnH-GUG, trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU) and 4 rRNAs (rrn4.5, rrn5, rrn16 and rrn23). Most of the PCG genes contained only one exon, while 18 genes contained one or two introns. Out of the 18 intron-containing genes, 10 PCG genes (atpF, ndhA, ndhB, rpoC1, petB, petD, rpl2, rpl16, rps12 and rps16) and 6 tRNAs (trnA-UGC, trnI-GAU, trnG-GCC, trnK-UUU, trnL-UAA and trnV-UAC) had a single intron, while two other genes (ycf3 and clpP) possessed two introns. The nucleotide composition was asymmetric (31.62% A, 18.42% C, 17.78% G, 32.18% T) with an overall AT content of 63.80%. The AT contents of the LSC, SSC, and IR regions were 65.99%, 70.40%, and 58.85%, respectively.
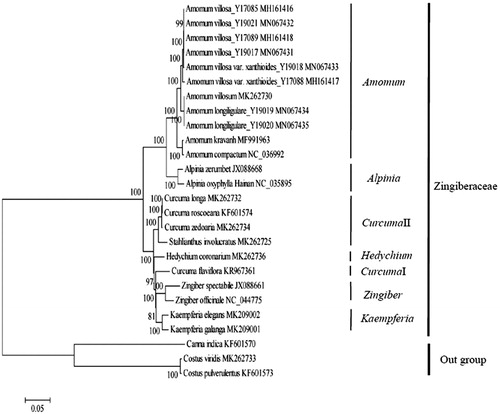
To obtain its phylogenetic position within the family Zingiberaceae, a molecular phylogenetic tree was constructed by using single nucleotide polymorphisms (SNPs) arrays from the available 26 chloroplast genomes using Costus viridis, Costus pulverulentus and Canna indica as outgroup taxa. The SNP arrays were obtained according to the previously described method (Li et al. Citation2019). For each chloroplast genome, all SNPs were connected in the same order to obtain a sequence in FASTA format. Multiple FASTA format sequences alignments were carried out using MEGA7 (Kumar et al. Citation2016). A maximum likelihood phylogenetic tree () was constructed using the SNPs from 26 chloroplast genomes alignment result with MEGA7 (Kumar et al. Citation2016). As shown in the phylogenetic tree (), C. zedoaria, Curcuma roscoeana, Curcuma longa and Stahlianthus involucratus clustered together in group CurcumaII within the family Zingiberaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Branney T. 2005. Hardy Gingers: including Hedychium, Roscoea and Zingiber. Galesburg, IL: Royal Horticultural Society/Timber Press, Inc.
- Dosoky NS, Setzer WN. 2018. Chemical composition and biological activities of essential oils of Curcuma speceies. Nutrients. 10(9):1196.
- Kumar S, Stecher G, Tamura K. 2016. MEGA 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li X, Hu Z, Lin X, Li Q, Gao H, Luo G, Chen S. 2012. High-throughput pyrosequencing of the complete chloroplast genome of Magnolia officinalis and its application in species identification. Yao Xue Xue Bao. 47(1):124–130.
- Li DM, Zhao CY, Liu XF. 2019. Complete chloroplast genome sequences of Kaempferia galanga and Kaempferia elegans: molecular structures and comparative analysis. Molecules. 24(3):474.
- Makabe H, Maru N, Kuwabara A, Kamo T, Hirota M. 2006. Anti-inflammatory sesquiterpenes from Curcuma zedoaria. Nat Prod Res. 20(7):680–685.
- Matsuda H, Tewtrakul S, Morikawa T, Nakamura A, Yoshikawa M. 2004. Anti-allergic principles from Thai zedoary: Structural requirements of curcuminoids for inhibition of degranulation and effect on the release of TNF-alpha and IL-4 in RBL-2H3 cells. Bioorg Med Chem. 12(22):5891–5898.
- Wilson B, Abraham G, Manju VS, Mathew M, Vimala B, Sundaresan S, Nambisan B. 2005. Antimicrobial activity of Curcuma zedoaria and Curcuma malabarica tubers. J Ethnopharmacol. 99(1):147–151.
- Zhou L, Zhang K, Li J, Cui X, Wang A, Huang S, Zheng S, Lu Y, Chen W. 2013. Inhibition of vascular endothelial growth factor-mediated angiogenesis involved in reproductive toxicity induced by sesquiterpenoids of Curcuma zedoaria in rats. Reprod Toxicol. 37:62–69.