Abstract
Forsythia suspensa is an important medicinal plant and native to China. In this article, the complete chloroplast genome of F. suspensa was studied and annotated. The chloroplast genome of F. suspensa is 156,404 bp in length, which has a large single-copy region (LSC) of 87,150 bp, a small single-copy region (SSC) of 17,812 bp, and two of inverted-repeat regions (IRs) of 25,716 bp. The overall nucleotide composition is: 30.8% (48,103 bp) of A, 31.4% (49,193 bp) of T, 19.2% (30,066 bp) of C and 18.6% (29,042 bp) of G, and the total content of the chloroplast genome A + T content of 62.2% and G + C content of 37.8%. The chloroplast genome of F. suspensa contains 133 genes, including 88 protein-coding genes (PCGs), 37 transfer RNA (tRNAs), and 8 ribosome RNA (rRNAs). The phylogenetic tree result shows that F. suspensa is closely related to F. intermedia in the phylogenetic relationship by the maximum-likelihood (ML) method.
Forsythia suspensa belongs to the Oleaceae family and native to China. From China, it (‘Yin-Qiao’ in Chinese) has been an important medicinal plant for many years, which is traditionally used for the treatment of inflammation, pyrexia, gonorrhea, diabetes, etc. (Ha et al. Citation2018). But, Zhejiang integrated hospital of traditional Chinese and Western medicine used three-dimensional printing technology to prepare multi-drug slow-release drug-loaded artificial bone, combined with Forsythia, which has a significant effect on sacroiliac joint tuberculosis (Ma et al. Citation2018). In order to further study the phylogenetic relationship of F. suspensa with the medicinal plant, we studied and annotated the complete chloroplast genome of F. suspensa, which will be useful in the clinical drug development of the herb plant and its valuable medicinal research in future.
The F. suspensa samples were collected from the herb market near Zhejiang Chinese Medical University located at Hangzhou, Zhejiang, China (119.89E, 30.09 N). The chloroplast genome DNA of F. suspensa was extracted from the fresh samples using the Plant Tissues Genomic DNA Extraction Kit (Solarbio, BJ, and CN) and stored in the Zhejiang Chinese Medical University (No. ZJCMU-002). The chloroplast genome DNA was purified and sequenced. Quality control was performed to remove low-quality reads and adapters using the FastQC (Andrews Citation2015). The chloroplast genome was assembled and annotated using the MitoZ software (Meng et al. Citation2019). The physical map of the chloroplast genome of F. suspensa was drawn using OGDRAW (Lohse et al. Citation2013).
The complete chloroplast genome of F. suspensa (GenBank with accession No. MK7534872) was 156,404 base pairs in length and had a typical quadripartite structure with a large single-copy region (LSC) of 87,160 bp, a small single-copy region (SSC) of 17,812 bp, and a pair of inverted-repeat regions (IRs) of 25,716 bp. The chloroplast genome of F. suspensa contains 133 genes, which included 88 protein-coding genes (PCGs), 37 transfer RNA genes (tRNAs), and 8 ribosomal RNA genes (rRNAs). In the IR region, 19 genes were found that included 8 PCGs (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, rps12 and ycf1), 7 tRNA (trnL-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG and trnN-GUU), and 4 rRNA (rRNA16, rRNA23, rRNA4.5 and rRNA5). The overall nucleotide composition was: 30.8% (48,103 bp) of A, 31.4% (49,193 bp) of T, 19.2% (30,066 bp) of C and 18.6% (29,042 bp) of G, with the total A + T content of 62.2% and G + C content of 37.8%.
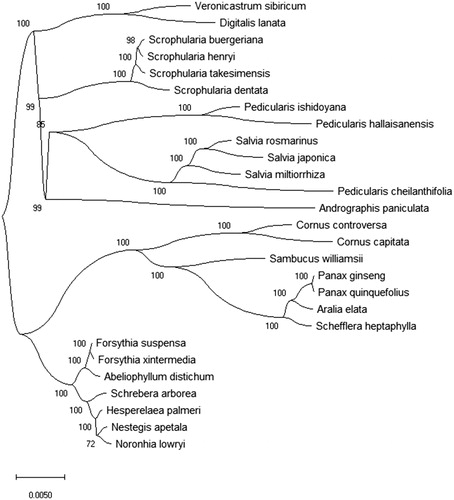
In this article, the maximum-likelihood (ML) method was used to analyse the phylogenetic relationship of 26 herb species plants with F. suspensa. The phylogenetic tree was reconstructed using the MEGA X (Kumar et al. Citation2018) with 2000 bootstrap values replicating at each node. All of the nodes were inferred with strong support by the ML methods. The phylogenetic tree was represented using the MEGA X and edited using the iTOL 4.0 (https://itol.embl.de/) (Letunic and Bork Citation2016). The phylogenetic tree result () showed that F. suspensa is closely related to Forsythia intermedia (GenBank No. MG255756.1). This study can be used for the clinical drug development of the herb and its medicinal valuable research in future.
Figure 1. The Maximum-Likelihood (ML) phylogenetic tree was constructed using 27 herb species plants chloroplast genomes sequences. ML bootstrap support value presented at each node. The NCBI GenBank accession number is: Abeliophyllum distichum KT274029, Andrographis paniculata KF150644, Aralia elata KT153023, Cornus capitata MG524998, Cornus controversa MG525004, Digitalis lanata KY085895, Forsythia xintermedia MG255756, Hesperelaea palmeri LN515489, Nestegis apetala MG255758, Noronhia lowryi MG255759, Panax ginseng KM088019, Panax quinquefolius KT028714, Pedicularis cheilanthifolia KY751712, Pedicularis hallaisanensis MG770330, Pedicularis ishidoyana KU170194, Salvia japonica KY646163, Salvia miltiorrhiza HF586694, Salvia rosmarinus KR232566, Sambucus williamsii KX510276, Schefflera heptaphylla KT748629, Schrebera arborea MG255767, Scrophularia buergeriana KP718626, Scrophularia dentata MF861202, Scrophularia henryi MF861203, Scrophularia takesimensis KM590983, Veronicastrum sibiricum KT724053.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. [accessed 2020 Feb 24]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Ha Y-H, Kim C, Choi K, Kim J-H. 2018. Molecular phylogeny and dating of Forsythieae (Oleaceae) provide insight into the Miocene history of Eurasian temperate shrubs. Front Plant Sci. 9:99.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581.
- Ma P F, Shi S Y, Fei J, Lai Z, Shi L F, Liu F. 2018. 3D printing of thoracic tuberculosis foci and vertebra model to guide the placement of pedicle screws. Zhejiang J Integrat Tradit Western Med. 28:302–306.
- Meng G L, Li Y Y, Yang C T, Liu S L. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
