Abstract
The complete mitochondrial genome of Cynoglossus roulei was determined in this study. The 16,565-bp-long genome is made of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and two noncoding regions. We detected the rearrangement of tRNA-Gln and control region in C. roulei. Additionally, tRNA-Gln gene was inverted from the L-strand position to the H-strand position. Phylogenetic analysis showed that C. roulei is most closely related to C. semilaevis.
Cynoglossus roulei (Pleuronectiformes: Cynoglossidae) an edible demersal fish found in China currently distributed across Hainan, Guangxi and Guangdong provinces to the south of Zhoushan Islands (Li and Wang Citation1995). In this study, we determined the complete mitochondrial genome of C. roulei and analyzed its phylogenetic position.
The specimen of C. roulei (Voucher No. ZJRA2012061721) was collected from Ruian, Zhejiang province. The protocol and data analysis method followed by Chen et al. (Citation2016). The complete mitochondrial genome of C. roulei (GenBank accession number: MN966658) is 16,565 bp in length, which contains 13 protein-coding genes, 22 tRNA genes, two rRNA genes, one control region (CR), and origin of light-strand replication(OL). Among these genes, ND6 and 8 tRNA genes are encoded on the Light-strand, while others are encoded on the Heavy-strand. Most protein-coding genes have typical initiation codons (ATG or GTG) and termination codons (TAA or TAG), except for the ND3 started with ATT, ND4 and COII terminated with T––. The size of 13 protein-coding genes ranged from 165 bp (ATP8) to 1854 bp (ND5), and 22 tRNA genes ranged from 66 bp (tRNA-Cys) to 75 bp (tRNA-Lys). The 12S rRNA (946 bp) and 16S rRNA genes (1683 bp) located between tRNA-Phe and tRNA-Leu(UUR) and separated by tRNA-Val. The Origin of light-strand is 33 bp long between tRNA-Asn and tRNA-Cys. The control region of the C. roulei mitogenome is 821 bp long and is located between ND1 and tRNA-Gln instead of the typical location between tRNA-Pro and tRNA-Phe. Furthermore, the tRNA-Gln gene was inverted from the L-strand position to the H-strand position. This result is consistent with the findings of previous studies where Pleuronectiformes was found to have large-scale gene rearrangements, including control region translocation, tRNA-Gln gene inversion and tRNA-Ile gene shuffling (Shi et al. Citation2013).
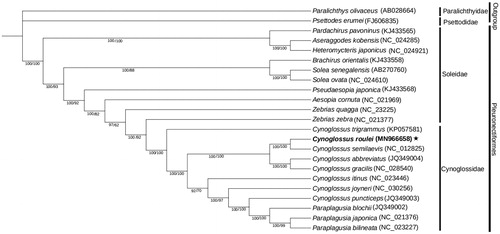
The phylogenetic tree was constructed based on 11 Cynoglossidae species and 10 Soleidae species with Psettodes erumei and Paralichthys olivaceus as the outgroups (), whose mitochondrial genomes were available in GenBank. Phylogenetic analyses were performed using the Bayesian analyses (BI) and Maximum-Likelihood (ML) methods in PhyloSuite (Zhang et al. Citation2020). Data were concatenated by two gblocks (first and second codons of 12 protein-coding genes, except ND6) and two rRNA genes. Cynoglossus roulei has a close phylogenetic relationship to C. semilaevis when compared with other Cynoglossus species (), then this clade was the sister group of C. abbreviatus + C. gracilis clade. All Paraplagusia species clustered together and are derived from Cynoglossus. The result shows mitochondrial genomic data are important for analyzing phylogenetic relationships in the Cynoglossinae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chen X, Ai WM, Xiang D, Shi XF, Pan LH. 2016. The complete mitochondrial genome of the wavyband sole Pseudaesopia japonica (Pleuronectiformes: Soleidae). Mitochondrial DNA. 27(1):420–421.
- Li SZ, Wang HM. 1995. Fauna sinica (Osteichthyes): Pleuronectiformes. Beijing (China): Science Press.
- Shi W, Dong XL, Wang ZM, Miao XG, Wang SY, Kong XY. 2013. Complete mitogenome sequences of four flatfishes (Pleuronectiformes) reveal a novel gene arrangement of L-strand coding genes. BMC Evol Biol. 13(1):173.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li W.X, Wang G.T. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.